Structure Function Group
DNA Polymerases Lambda (λ) and Mu (μ) are members of the X-Family, and are primarily involved in DNA repair. Both polymerases function in repair of DNA double strand breaks (DSBs), a particularly toxic type of lesion that can result in chromosomal rearrangements or cell death if not corrected. In nonreplicating cells, these breaks are commonly repaired by a process known as nonhomologous end-joining (NHEJ). DSBs occur in a programmed manner during V(D)J recombination, a process that leads to maturation of immunoglobulin genes. However, DSBs also result from exposure to gamma-radiation, reactive oxygen species, and exogenous environmental agents.
Pols λ and μ both function in NHEJ, though each polymerase is recruited to specific types of broken DNA ends. Pol λ has a strict requirement for a paired primer terminus. In contrast, Pol μ is the only known eukaryotic polymerase that can act on broken DNA ends with no sequence complementarity at the site of the break, i.e. synpases involving an unpaired primer terminus.
Our lab employs X-ray crystallography to understand the structural determinants of DNA binding, specificity and catalysis by these polymerases. In support of the Kunkel lab, we have solved crystal structures in complex with a variety of DNA substrates. Nonhydrolyzable analogs of naturally occurring nucleotides are used to capture catalytically relevant complexes on the cusp of the enzymatic reaction. Once the crystals have formed, we can remove such analogs, and replace them with the catalytically-competent nucleotide, which allows the reaction to run to completion in the crystal.
Our structures have revealed that there is a gradient of protein and DNA substrate movements that are essential for efficient catalysis by the Family X polymerases. Pol β, with the strictest substrate requirements, undergoes large-scale motions of protein subdomains, DNA substrate, and active site side chains during catalysis. Pol λ, with a slightly more flexible DNA substrate specificity, forgoes global protein subdomain motions, but uses more subtle adjustments of DNA template and active site side chains to achieve optimal positioning. By contrast, Pol μ functions as a rigid scaffold, and having few requirements for DNA template strand structure. Rigidity of the protein appears to allow for flexibility of template strand conformation or accommodation of distortions. Additionally, our structures reveal that, while most gap-filling polymerases use the upstream side of the gap (closest to the 3ʹ-OH on the primer terminus) to direct synthesis, Pol μ instead ‘skips ahead’, using the downstream end of the gap (closest to the 5ʹ-phosphate) as a guide. This ‘skip ahead’ mechanism appears to be a unique to Pol μ and is consistent with the reported behavior of this enzyme in vivo.
Recently we have been able to obtain crystal structures of Pol µ engaged in binding a DSB substrate and proceeded to turn the reaction over in crystallo, supporting the binding as being catalytically-relevant (1). These structures reveal little difference between how Pol µ engages single-strand versus complementary double-strand breaks, suggesting its rigid scaffold is important to its ability to fill DSBs. In addition, we have determined the first crystal structures of the catalytic domain of the ligase responsible for annealing these breaks, human LigIV, in complex with DNA and two distinct adenylation states of the catalytic pathway (2). These structural studies provide insight into how LigIV engages DNA substrates, the conformational changes associated with binding, the roles specific residues play in this process, and suggest how certain mutations may contribute to disease.
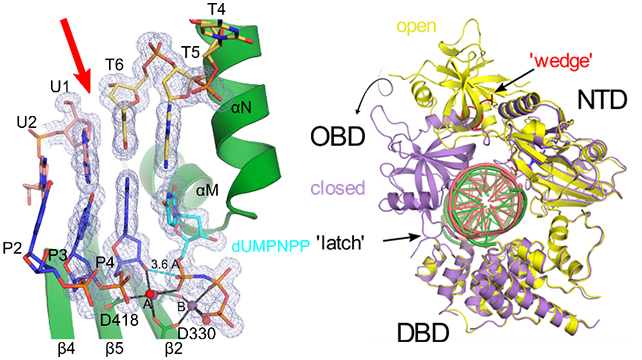
Left, crystal structure of Polμ (green) bound to a complementary double-strand break (DSB, break site indicated by red arrow) DNA substrate (upstream template and primer strands in pink and blue, downstream template strand in yellow, incoming dUMPNPP in cyan, Mg2+ ions and water molecules as purple and red spheres). 2Fo-Fc (gray mesh) is contoured at 1σ. Right, global structure of hLigIV in the open protein-adenylate (yellow) and closed DNA-adenylate (purple) states, with DNA bound (pink and green in the open and closed states, respectively). LigIV performs the final nick sealing step of NHEJ.
References
- Kaminski, A. M., Pryor, J. M., Ramsden, D. A., Kunkel, T. A., Pedersen, L. C., and Bebenek, K. (2020) Structural snapshots of human DNA polymerase mu engaged on a DNA double-strand break. Nat Commun 11, 4784
- Kaminski, A. M., Tumbale, P. P., Schellenberg, M. J., Williams, R. S., Williams, J. G., Kunkel, T. A., Pedersen, L. C., and Bebenek, K. (2018) Structures of DNA-bound human ligase IV catalytic core reveal insights into substrate binding and catalysis. Nat Commun 9, 2642


