About PSPE
PSPE is a computational tool specifically designed for simulating evolution of non-coding DNA sequences, in particular promoter sequences. PSPE, like other simulation tools, can use many different DNA substitution models and Insertion/Deletion models to simulate sequence evolution. What makes PSPE unique is that PSPE can perform sequence evolution under a set of user-specified functional constraints, e.g., GC contents, functional site locations, and distances between functional sites, which makes simulation more realistic. In addition, PSPE can, using a Poisson process for branching and a Exponential model for branch length, generate a random evolutionary tree of different topologies. The outputs of PSPE include simulated sequences, the alignment of sequences, and the location map of functional sites in each sequence. PSPE is a useful tool for studying evolution mechanisms and for evaluating alignment tools as well.
Availability and Citation
PSPE is freely available to public and can be downloaded at the following link.
PSPE for both 32 and 64 bits Linux systems (1MB)
Please use the following command to extract files from the compressed file: tar xvfj PSPE-Linu-Bin.tar.bz2, then follow the instructions in the README file for installation and usage.
Data
TFBS PWM for evolution simulation (359B)
Supplementary Results
Additional results on MSA tool evaluation (437KB)
Note: some above files for download are in *.tar.gz format (use command 'tar xfz afile.tar.gz' to extract files). If you would like to have other data or programs used in our paper, please feel free to contact me.
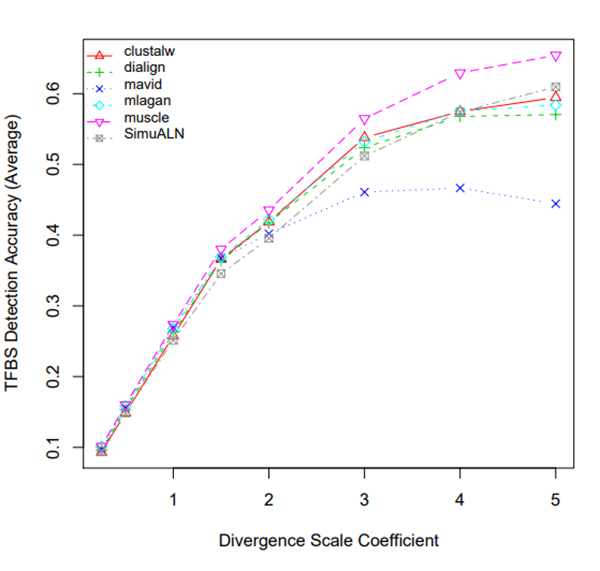
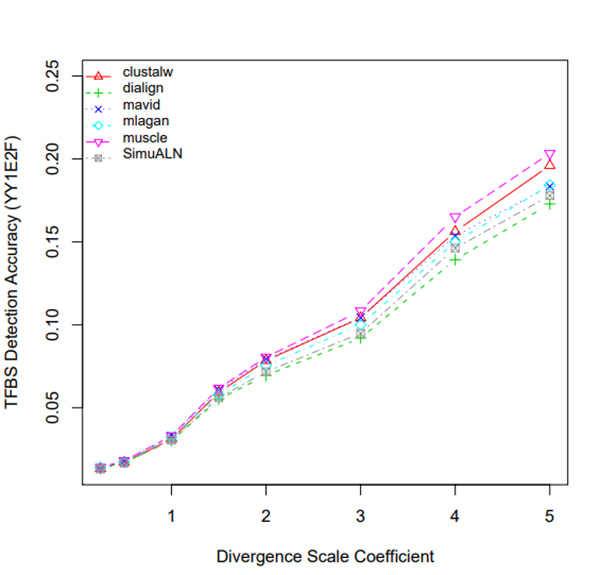
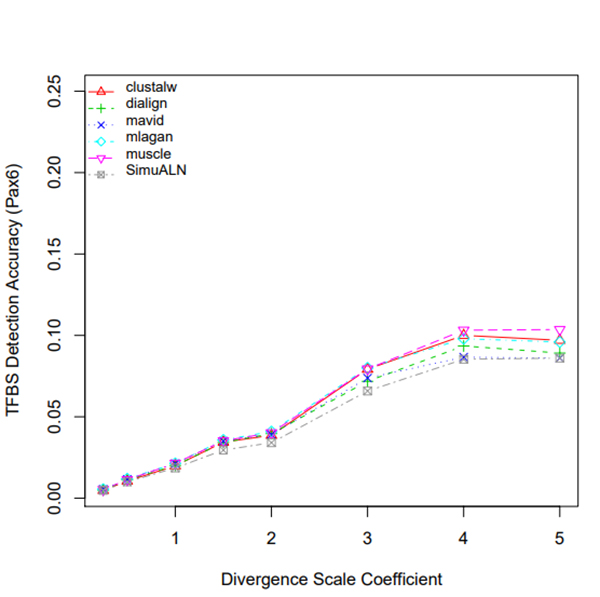
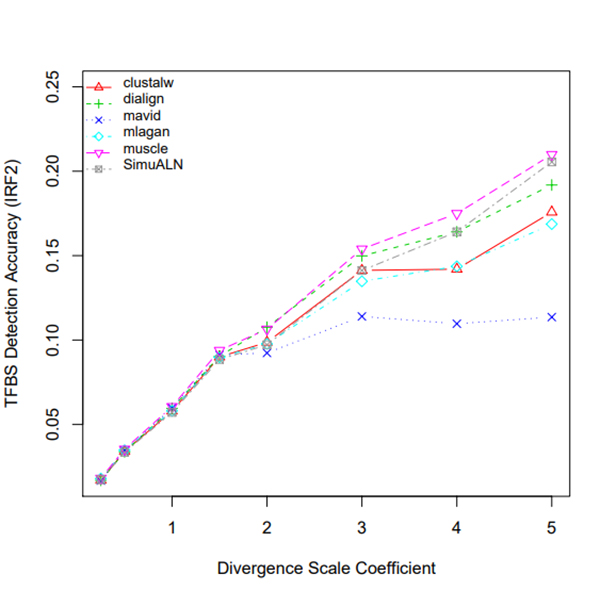
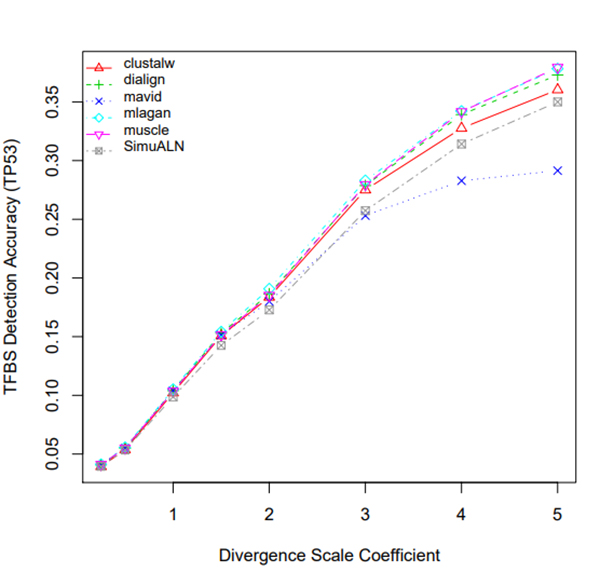
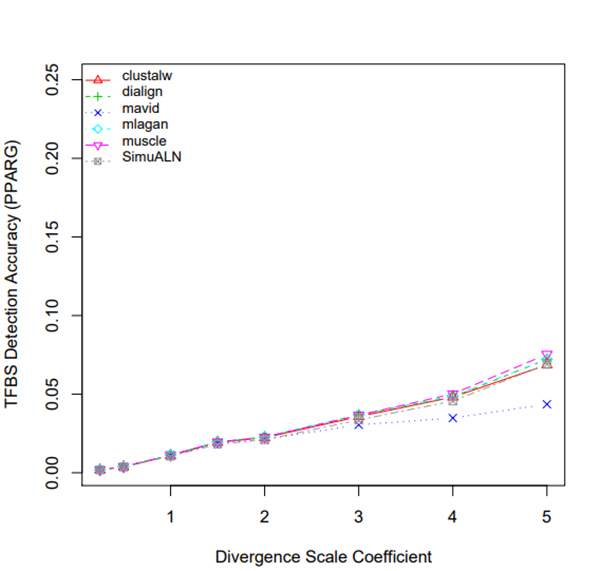
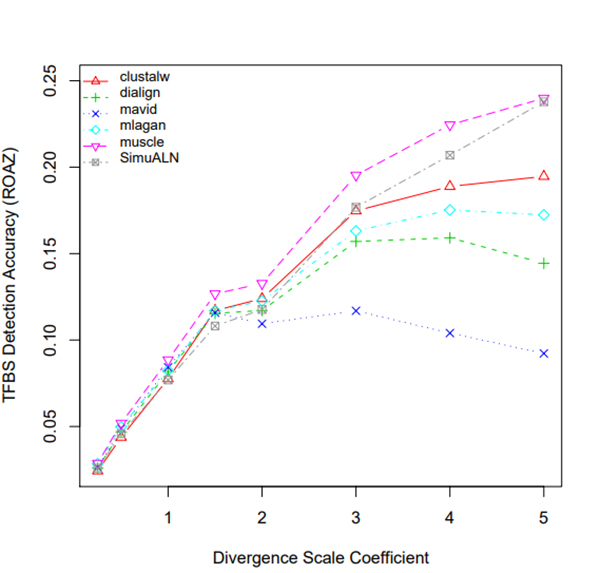
R Graphics Output
TFBS Detection Accuracy of Turnover Sites in Aligning Five Mammal Species