About ACANA
ACANA is an accurate and consistent alignment tool for DNA sequences. ACANA is specifically designed for aligning sequences that share only some moderately conserved regions and/or have a high frequency of long insertions or deletions. It attempts to combine the best of local and global alignments algorithms in searching for evolutionarily related regions of sequences in order to achieve the best alignment. ACANA is also robust to the small changes of alignment parameters, particularly the gap extension score. As an accurate alignment tool, ACANA is particularly useful in comparative sequence analysis for identifying conserved functional regulatory elements.
Availability and Citation
ACANA is freely available for download at the following links.
64-bit systems
- ACANA for 64-bit MacOS X (Intel CPU) - ACANA_MacOSX_x64.tar.gz (125KB)
- ACANA for 64-bit Linux - ACANA_Linux_x64.tgz (3MB)
- ACANA for 64-bit Windows - ACANA_MsWindows64.zip (2MB)
32-bit systems
- ACANA for 32-bit MacOS X - ACANA_MacOS_X_Darwin.tar.gz (492KB)
- ACANA for 32-bit Linux - ACANA_Linux_x32.tgz (3MB)
- ACANA for 32-bit Windows - ACANA_MsWindows32.zip (653KB)
Note: Please use a 64-bit version for aligning long sequences as a 32-bit program can use up to 4GB memory only. To uncompress files, please use one of the following commands :
1) tar xvfz *.tar.gz
OR
2) unzip *.zip
then follow the instructions in the README file for installation and usage.
ACANA should be cited as
Weichun Huang, David M. Umbach, and Leping Li, Accurate anchoring alignment of divergent sequences. Bioinformatics 22:29-34, Jan 1 2006 DOI:10.1093/bioinformatics/bti772i [Abstract] [Full Text] [Supplementary-HTML] [Supplementary PDF] (251KB)
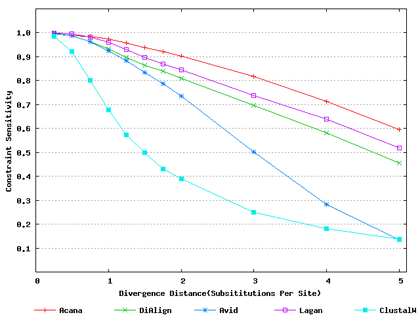
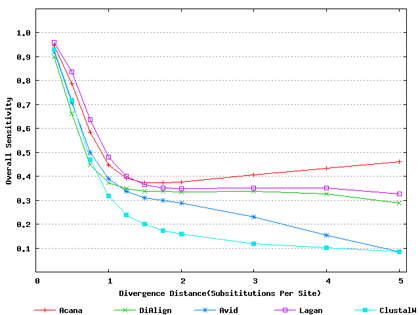
Test Data and Results
Simulated Data: The benchmark data of non-coding DNA sequences were from Pollard, Bergman, Stoye, Celniker & Eisen (2004) BMC Bioinformatics 5(1):6.


