Epigenomic Transcriptional Regulation

Richard Woychik, Ph.D.
Research Summary
Richard Woychik, Ph.D., is Director of NIEHS & NTP and head of the Mammalian Genome Group. The group is broadly interested in understanding the role of epigenomic transcriptional regulation in an organism’s response to environmental exposures. It is specifically interested in studying the role of repetitive elements (REs) in these processes.
All mammalian genomes are comprised of a large assortment of different classes repetitive elements (REs) that include LINE’s, intracisternal A particles (IAPs), VL30 elements, endogenous retroviral elements (ERVs), SINEs, transposons, and various other types of repetitive elements. These REs are distributed across the entire genome and are highly abundant. In fact, it has been estimated that REs comprise more than 50 percent of a typical mammalian genome. Many of these elements have powerful regulatory elements and exhibit complex patterns of transcriptional expression throughout mammalian development.
Transcription of repetitive elements has been shown to influence the expression of adjacent genes. Since there are nearly 5 million RE loci across the mammalian genome, the group is currently working to determine how many genes have patterns of expression that are influenced by adjacent REs. The group is also examining the role of mitochondrial metabolism in the epigenetic regulation of genes, including REs, within the nuclear genome.
The group utilizes state-of-the-art NextGen genome analysis technologies, bioinformatics, and molecular and genetic methods. Group members actively work to foster interdisciplinary exchanges with postdoctoral fellows integrating bioinformatics, statistics, genetics, and biochemical and molecular biology approaches.
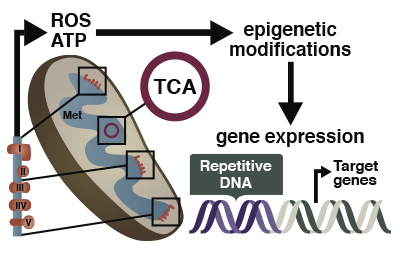
Major areas of research:
- Environmental exposures, biology of REs, epigenomic regulation of gene expression, and transcriptome analysis
- Crosstalk between mitochondrial function and the epigenome
Current projects:
- Transcriptome analysis of biological samples treated with specific environmental agents
- Understanding the impact of sequential loss of mitochondrial DNA and oxidative phosphorylation function on the epigenome
- Development of in silico methods to detect fusion transcripts between REs and DNA coding regions
Woychik completed his B.S. and M.S. at the University of Wisconsin, Madison, and earned his Ph.D. in molecular biology at Case Western Reserve University in 1984. He received his postdoctoral training in the laboratory of Philip Leder, M.D., at Harvard Medical School with fellowship support from the Jane Coffin Childs Memorial Fund and from the Howard Hughes Medical Institute.


