Introduction
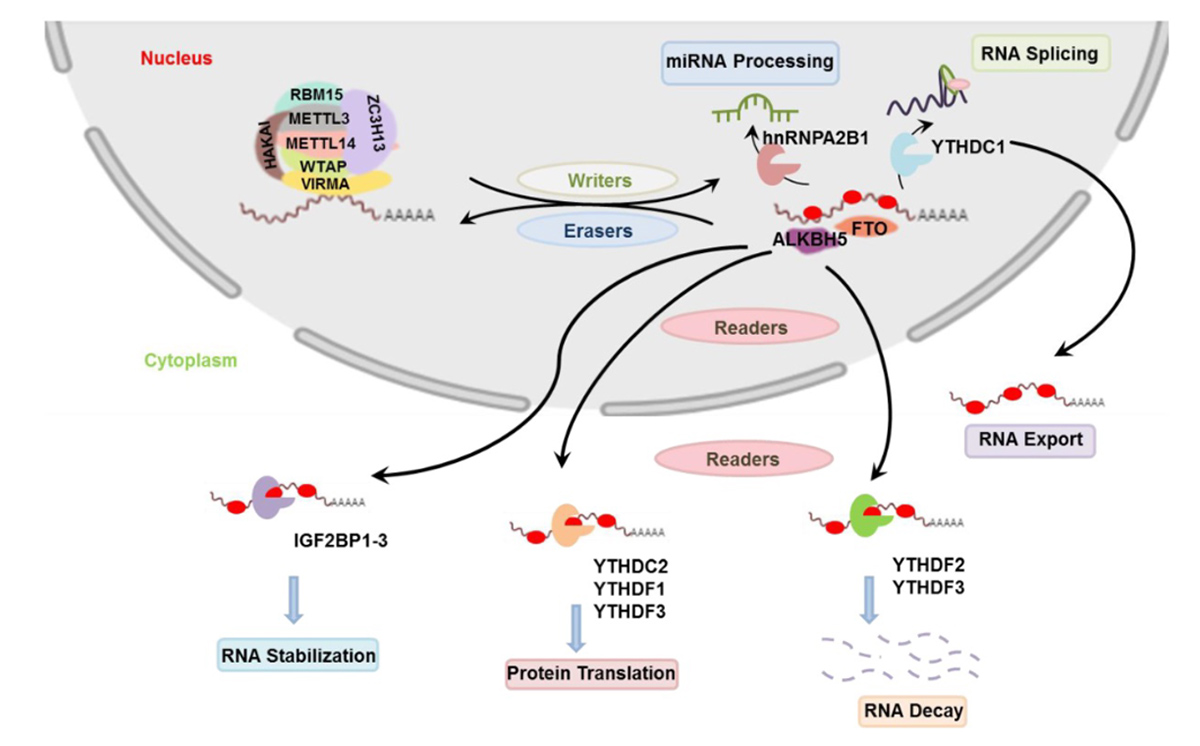
Chemical modifications of protein, DNA, and RNA molecules play critical roles in regulating gene expression. Emerging evidence suggests post-transcriptional RNA modifications have major roles in multiple basic biological processes. Epitranscriptomics (also called RNA epigenetics) can be defined as all RNA modifications that exist within a cell. Just as epigenetic modifications regulate gene expression by modulating DNA accessibility, epitranscriptomic changes regulate gene expression by affecting RNA stability, localization, and processing. These chemical modifications are controlled by proteins that function as readers, writers, and erasers (RWEs). Writers and erasers can add or remove specific modifications, respectively, while readers interpret and process them. Studies in yeast, fruit flies, rodents and human models demonstrate that stressors can induce RNA modifications, with specific reprogramming of some regulatory RNAs.
What NIEHS Is Doing
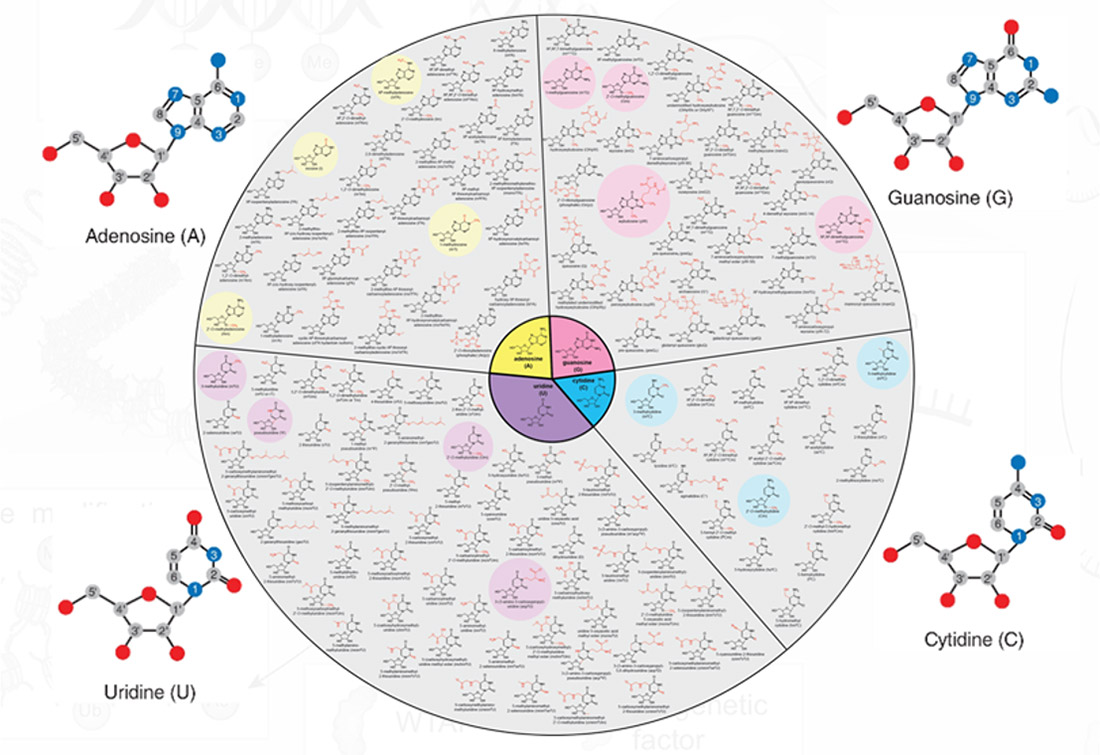
The NIEHS environmental epitranscriptomics portfolio supports research focused on how environmental exposures affect the epitranscriptome and related health impacts. So far, over 170 RNA base modifications have been identified that have far-reaching impacts on molecular pathways associated with the development and progression of adverse health outcomes. Researchers are using a variety of methods, including cell cultures, animal models and population-based approaches to determine how environmental toxicants impact RNA biology and human health. Changes to the epitranscriptome are also being studied as potential biomarkers of exposure and exposure-induced pathologies.
The portfolio currently has approximately 40 active grants that examine a number of exposures including metals (As, Ni), phosphate, UVB, vinyl chloride, high fat diets (HFDs), and agents that induce reactive oxygen species (ROS). These grants cover a range of health outcomes such as transgenerational metabolic outcomes, lung and cardiovascular disease, cancers, neurodevelopmental and neurodegenerative disorders. There are currently over 100 diseases that are associated with aberrant epitranscriptomic processes. How environmental exposures could impact epitranscriptomics and related disorders is an ongoing area of research.
As our ability to sequence and analyze the epitranscriptome evolves, so too will the research that NIEHS supports. NIEHS-funded environmental epitranscriptomics research could enhance our understanding of how environmental factors influence human health and change how we diagnose and treat disease.
NIEHS Supported Research
Stress and the Epitranscriptome
The ability of the epitranscriptome to respond to stress is vital for health and survival. One of the most prevalent RNA modifications, m6A, regulates mRNA stability, processing, localization, and translation efficiency depending on its location within the mRNA strand. In response to oxidative stress, the writer proteins METTL3/METTL14 deposit m6A, inducing upregulation of antioxidant genes. DNA damage resulting from UV irradiation also leads to m6A being recruited to the damaged sites and facilitating repair mechanisms. Other reader and writer proteins can also regulate protein translation and lipid metabolism in response to stress.
Exposures and Disease
Environmental exposures can cause changes in the epitranscriptome that contribute to disease pathology, such as liver disease and cancer. METTL3, METTL14, and FTO are writers that deposit m6A on RNA molecules. Overexpression occurs often in cancerous tissues and is associated with worse outcomes in cancer patients. RWEs and m6A dysregulation can be driven by toxic metal exposures such as arsenic, hexavalent chromium, and cadmium. Aberrant m6A modifications play a key role in carcinogenesis and the adverse health outcomes caused by these exposures. METTL3 and FTO have increased expression in fatty liver disease and drive increased m6A profiles in genes important for lipid metabolism. These proteins are impacted by a high fat diet and other exposures, such as vinyl chloride and polychlorinated biphenyls.
Transfer RNA (tRNA) plays a key role in protein translation and is heavily modified. These modifications affect tRNA structure, stability, and function, leading to significant effects on protein translation. Mutations in the enzymes that regulate tRNA modifications can cause multiple types of disorders such as cancer, neurological disorders, and mitochondrial diseases. Of the 75 proteins that have been found to regulate tRNA modifications, 54 have been associated with tRNA modopathies.
Epitranscriptomics and Development
METTL3 is critical during early development as m6A modifications are critical for regulating maternally deposited mRNA in early development. m6A readers and writers are involved in neurodevelopment and important for processes such as neuron differentiation, axon guidance, and astrocyte proliferation. How early exposures might impact the epitranscriptome and drive developmental defects is an ongoing area of study.
Similar to DNA methylation, RNA modifications can also be passed down from parent to offspring. Recent studies have shown that small RNAs in sperm can be passed from one generation to the next. Environmental exposures can modify sperm RNA which contribute to offspring phenotypes in a process referred to as transgenerational inheritance.
Grantees
| Project Title | Principal Investigator | Institution | Grant Number |
|---|---|---|---|
| ALKBH5 and Nickel-Induced Lung Carcinogenesis | Hong Sun, Ph.D. | New York University School of Medicine | R21ES034811-02 |
| Chemical Biology of DNA and RNA Alkylation | Yinsheng Wang, Ph.D. | University of California Riverside | R35ES031707 |
| Decoding the Signature of Sperm RNA & RNA Modification of Environmental Stressors on the Intergenerational Transmission of Metabolic Phenotypes | Qi Chen, Ph.D. | University of Utah | R01ES032024-05 |
| Determining the Role of RNA abasic Sites in Gene Regulation | Vivian Cheung, Ph.D. | Brown University | R21ES034919-02 |
| Dysregulations of Functional RNA Modifications and Hexavalent Chromium Lung Carcinogenesis | Chengfeng Yang, Ph.D. | State University of New York at Stony Brook | R01ES032787-04 |
| Epitranscriptomic Mechanism of Environmental Stress Response and Tumorigenesis | Yu-Ying He, Ph.D. | University of Chicago | R35ES031693-02 |
| Functional Alterations of the Dihydrouridine Landscape in Response to Environmental Stress | Wendy Gilbert, Ph.D. | Yale University | R21ES031525-02 |
| Functional RNA Modifications, Micronutrient Exposure, Developmental Disabilities | Hehuang Xie, Ph.D. | Virginia Polytechnic Institute and State University | R01ES031521-05 |
| Heavy Metal-Stimulated Signal Transduction: New Metal-Regulatory and -Responsive Mechanisms | Matthew Ross, Ph.D. | University of Chicago | K99ES034084-01A1 |
| Imprinted Gene Regulation by in utero Lead Exposure in Mice | Bambarendage Pinithi Perera, Ph.D. | University of Michigan at Ann Arbor | K01ES035064-01A1 |
| Mechanisms of Transgenerational Epigenetic Inheritance | Victor Corces, Ph.D. | Emory University | R01ES027859-07 |
| METTL3 in Chromium-Induced Angiogenesis and Carcinogenesis | Steven McMahon, Ph.D. | Thomas Jefferson University | R01ES033197-03 |
| MGMT Down-Regulation in the Carcinogenicity of Hexavalent Chromium | Zhishan Wang, Ph.D | State University of New York Stony Brook | R01ES029496-05 |
| Molecular Mechanisms Underlying Metabolic Reprogramming by Paternal Benzene Exposure | Heidi Lempradl, Ph.D. | Van Andel Research Institute | R56ES034765-01A1 |
| Modulation of RNA Binding Proteins in Xenobiotic-Induced Hepatotoxicity | Yogesh Saini, Ph.D. | North Carolina State University | R01ES033709-02 |
| N6-Methyladenosine (m6A) Interplays with RNA and DNA Damage to Regulate DNA Repair | Yuan Liu, M.D., Ph.D. | Florida International University | R03ES035200-02 |
| Nitric Oxide as a Novel Regulator of Alternative Splicing | Joseph Schindler, M.D., Ph.D. | Case Western Reserve University | F30ES035247-02 |
| Novel Epitranscriptomic Mechanisms in Metal Neurotoxicity | Anumantha Gounder Kanthasamy, Ph.D. | Iowa State University | 1R01ES036241 |
| Oxidative Stress and RNA Methylation | Ricardo Aguiar, Ph.D. | University of Texas Health Science Center | R01ES031522-05 |
| Prenatal Traffic-Related Air Pollutants, Placental Epitranscriptomics, and Child Cognition | Julie Herbstman, Ph.D. | Columbia University Health Sciences | R01ES032818-03 |
| Pho-m6A Assay: A Phosphoselective Method to Quantify Dynamics of m6A in mRNA | Qiuying Chen, Ph.D. | Weill Medical College of Cornell University | R21ES032347-02 |
| RNA Modifications by Paternal Exposure to Arsenic and Intergenerational Effects on Sperm Quality | Shuk-Mei Ho, Ph.D. | University of Arkansas for Medical Sciences | R01ES032675-03 |
| Role of m6A RNA Modifications in AHR-Mediated Developmental Toxicity | Neelakanteswar Aluru, Ph.D. | Woods Hole Oceanographic Institution | R21ES035153-01 |
| Role of PXR in EDC-Induced Cardiovascular Disease | Changcheng Zhou, Ph.D. | University of California, Riverside | R35ES035015-02 |
| Systems-Wide Analysis of Oxidative Stress-Responsive m6A Epitranscriptome | Yi-Lan Weng, Ph.D. | The Methodist Hospital Research Institute | R01ES031511-04 |
| The Epitranscriptome as a Novel Mechanism of Arsenic-Induced Diabetes | Ana Navas-Acien, Ph.D. | Columbia University Health Sciences | R01ES032638-04 |
| The Placental Epitranscriptome as a Novel Mechanism Behind Prenatal Metal Mixture Exposures and Child Growth and Development | Allison Kupsco, Ph.D. | Columbia University Health Sciences | R01ES035908-01 |
| The Role of m6A-RNA Methylation in Memory Formation and Recall and its Modulation and Influence on Long-Term Outcomes as a Consequence of Early Life Lead Exposure | Jay Schneider, Ph.D. | Thomas Jefferson University | R01ES034077-02 |
| Translational Regulation During Cigarette Smoking-Induced Reprogramming of the tRNA Epitranscriptome, in vitro and in a Mouse Smoking Model | Thomas Begley, Ph.D. | State University of New York At Albany | R01ES031529-05 |
| Translational Regulation in Exposure Biology - Xenobiotic-Induced Reprograming of tRNA Modifications and Selective Translation of Codon-Biased Response Genes in Rat and Human Models | Thomas Begley, Ph.D. | State University of New York At Albany | R01ES026856-09 |
| Understanding mRNA Condensation and Its Role in Translational Control During Stress | Hendrik Glauninger | University of Chicago | F30ES032665-04 |
| Using Riboglow to Define RNP Interactions in Response to Environmental Stress | Erin Richards | University of Colorado Boulder | F31ES033919-03 |


