| Term | Definition(s) | Examples | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | An explanatory or critical comment, or other in-context information (e.g., pattern, motif, link), that has been associated with data or other types of information. [Source: NCIt C44272] | A GO annotation is a statement about the function of a particular gene. Annotations associate a gene/gene product with a GO term. | ||||||||||||||||
| Common data element (CDE) See also Data Element | CDEs are standardized, narrowly defined questions that pair with a set of specific allowable responses. They can be used across different sites, research studies, or clinical trials to ensure consistent data collection. [Source: CDE Tutorial] | The NIH Common Data Elements Repository offers access to CDEs recommended or required by NIH Institutes and others. The PhenX Toolkit offers standard protocols. | ||||||||||||||||
| Controlled vocabulary | A controlled vocabulary (CV), also called an authority file or term list, is an authoritative set of terms selected and defined based on the requirements set out by the user group. A CV is used to ensure consistent indexing (human or automated) or description of data or information. Controlled vocabularies do not necessarily have any structure or relationships between terms within the list. [Source: NCIt C48697 and About Taxonomies & Controlled Vocabularies] | Some definitions of controlled vocabulary are more expansive and include taxonomy, thesaurus, ontology, etc. For our purposes, it is being considered only as a term list typically encountered as drop-down pick list, index list of terms, tagging codes, etc. | ||||||||||||||||
| Data curation | A managed process, throughout the data lifecycle, by which data and data collections are cleaned, documented, standardized, formatted and inter-related. Such processes ensure the value of the data is fit for purpose, preserved over time, and available for discovery and reuse. A second meaning of the phrase is used in the context of extracting information from research articles and storing that information in a database. [Source: Wikipedia] | The Data Curation Network provides a useful checklist. | ||||||||||||||||
| Data dictionary | A collection of descriptions of the data objects or items in a dataset. A data dictionary is used to catalog and communicate the structure and content of data and provides meaningful descriptions for individually named data objects. A data dictionary typically includes:
Shared dictionaries ensure that the meaning, relevance, and quality of data elements are the same for all users. Data dictionaries also provide information needed by those who build systems and applications that support the data. |
Additional items include description, and required values, among others. | ||||||||||||||||
| Data elements | Information that describes a piece of data to be collected in a study. The description includes a data element name, definition, permissible values, and other attributes. [Source: CDE Glossary and NCIt C41002] | For example, patient information contains the data element “name” and “address.” Even address can be composed of several additional data elements; e.g. “street address”, “city”, “postal code”, etc. | ||||||||||||||||
Data harmonization | Data harmonization is an extension of data integration. The harmonization process combines data from different sources and reorganizes it according to a single schema to provide users with a comparable view of data from different studies. Data is combined by either identifying equivalent data elements between the sources or by developing unequivocable transformations between the elements, to create a view of the unified data. In some cases, transformations can lead to loss of information or subtle changes in meaning within the unified view. In the context of epidemiology: Making data from different sources comparable. The processes involved in producing inferentially equivalent data. | Learn more about HHEAR’s data harmonization, NCI’s Quest for Harmonized Data and role of data harmonization in a molecularly driven health system. | ||||||||||||||||
Data integration | The practice of consolidating data from disparate sources into a single dataset with the goal of providing a unified, single view of the data. Combining diverse datasets from disparate sources into one unified dataset or database. Data are accessed and extracted, moved, validated, cleaned, transformed, and loaded. | Repositories integrate data by bringing disparate sources and collating them in a single database to improve findability. | ||||||||||||||||
| Data model | A model that specifies the structure or schema of a dataset. A data model can be thought of as a diagram or flowchart that illustrates the relationships between data. The model provides a documented description of the data and thus is an instance of metadata. It is a logical, relational data model showing an organized dataset as a collection of tables with entity, attributes and relations. | Learn more from an example data model from NCI’s Genomic Data Commons Data Model. 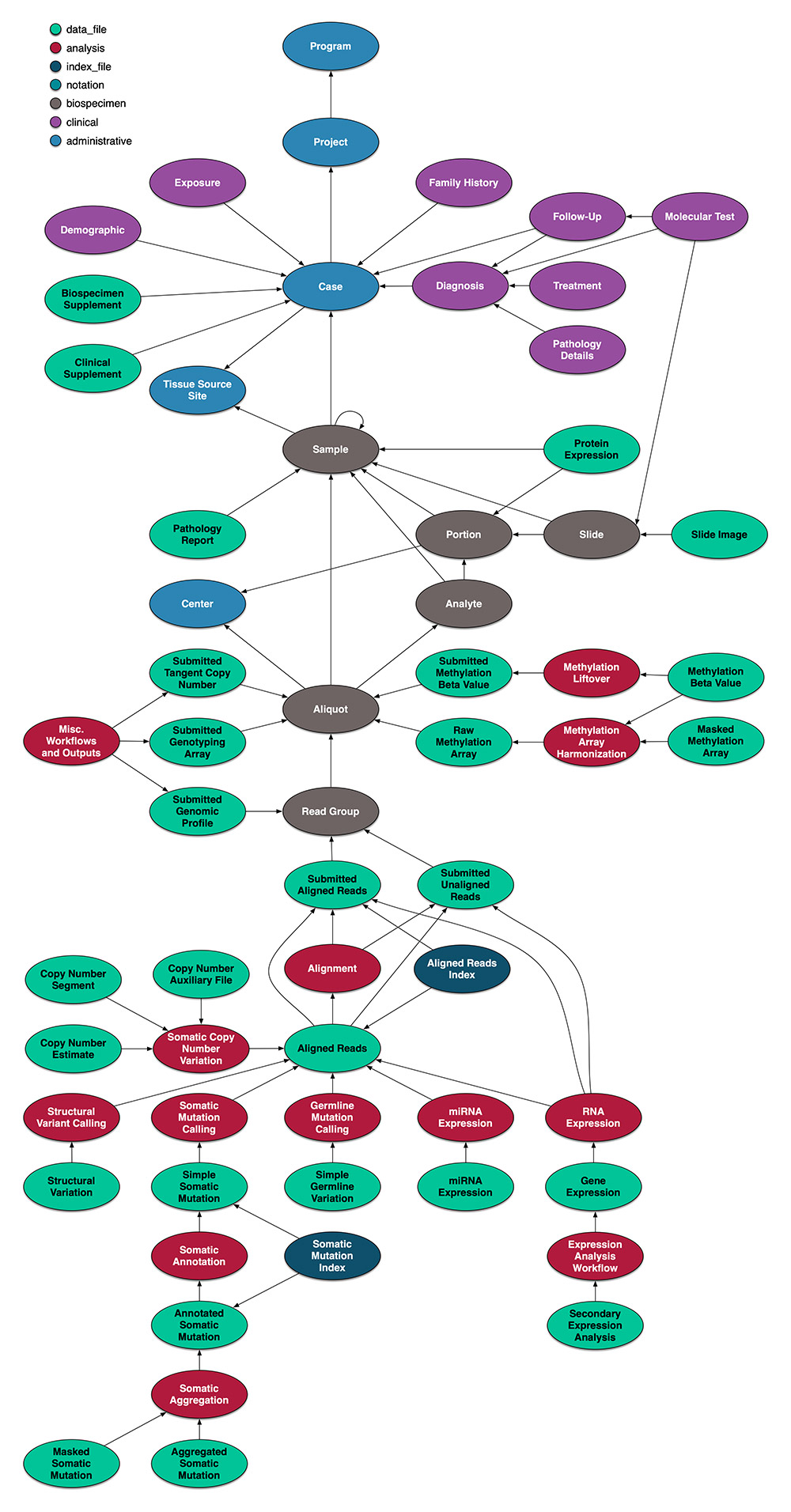
| ||||||||||||||||
| Data standards | Data standards are documented agreements on representation, format, definition, structuring, manipulation, use, and management of data. Data standards are needed for data to be presented and exchanged. [Source: EPA] | Identify domain-specific standards, models, reporting guidelines, and schemas FAIRsharing, including exploring the FAIR Cookbook tool. | ||||||||||||||||
| Harmonized language | A harmonized language combines multiple languages into a single comparable view building from the components of each language. [Source: Modified from ICPSR] | An example of harmonizing language could be if researchers have used a variety of beverage terms, such as cola, pop, soda, and soft drink. To integrate the data from studies using those different terms, each is matched to the harmonized term of carbonated beverage. | ||||||||||||||||
| Interoperability | Interoperability refers to the ability of two or more systems or components to exchange information and to use the information that has been exchanged. There are four types of issues that may impede interoperability:
Common language interoperability is a requirement to enable machine computable logic, inferencing, knowledge discovery, and data federation between information systems. | |||||||||||||||||
| Knowledge base | In general, a knowledge base is a database that holds statements about our knowledge in a particular domain instead of actual data points. More specifically, biomedical knowledgebases have the primary function to extract, accumulate, organize, annotate, and link growing bodies of information related to core datasets, in compliance with the FAIR Data Principles. | Database: Organism X was exposed to agent Y at latitude/longitude on date/time. Knowledgebase: Organism W resides near manufacturer X with emissions discharge Y leading to potential health outcomes Z. Comparative Toxicogenomics Database (CTD) is an example of a knowledgebase. It includes manually curated information from published literature on chemical–gene/protein-disease relationships with functional and pathway data to aid in development of hypotheses about the mechanisms underlying environmentally influenced diseases. | ||||||||||||||||
Knowledge graph | A method for representing knowledge as entities (nodes) and the relationship between them (edges) in a way that enables large-scale computing to take advantage of our knowledge of those relationships and make inferences of connections. [Source: based on An Introduction to Knowledge Graphs] | See knowledge graph example. | ||||||||||||||||
Knowledge organization | A term applied to all types of schemes (controlled vocabulary, taxonomy, ontology, etc.) used to organize, describe, represent, and manage a set of information. [Source: ISKO] | See the knowledge organization graphic. | ||||||||||||||||
Knowledge representation | A field of artificial intelligence that is concerned with presenting real-world information in a form that the computer can 'understand' and use to 'solve' real-life problems or 'handle' real-life tasks. [Source: Fingent] | |||||||||||||||||
| Metadata | Metadata is often called data about data or information about information. It ensures that the context for how your data was created, analyzed, and stored, is clear, detailed and therefore, more usable and reusable in the future. Metadata can be descriptive, administrative, or technical in nature. Metadata are structured, descriptive information of primary data and answer the five W-questions: What has been measured, by Whom, When, Where, and Why? | An experimental study may contain the following types of metadata:
| ||||||||||||||||
| Metadata standard | A standard that specifies what types of metadata should be collected and how for any given data, what format the metadata should be in, what units and terms should be used, and the file format the metadata should be in. [Source: adapted from Digital Curation Centre] | A few examples of metadata standards include: Cancer Data Standards Registry (caDSR), Crystallographic Information Framework. | ||||||||||||||||
| Minimum information standards | A specification of a minimum amount of information needed to reproduce or fully interpret a scientific result. The standard is typically composed of two parts: a table or checklist of reporting requirements and a data format. [Source: Ontobee and Wikipedia] | Numerous research methods use minimum information standards; e.g., MIATE (in vivo animal toxicology), MIAME (gene expression), MIBBI (biological and biomedical investigations. Find more at FAIRsharing. | ||||||||||||||||
Ontology | A formal representation of a body of knowledge within a given domain. An ontology is a controlled vocabulary of well-defined terms with specified relationships between them capable of interpretation by both humans and computers. Ontologies usually consist of a set of classes (or terms or concepts) with relations that operate between them. Ontologies are used to provide the underlying common language structure for knowledge graphs to ensure shared meaning and understanding of the data both by humans and machine. [Source: About Taxonomies & Controlled Vocabularies and Ontotext] | Human Health Exposure and Analysis Resource (HHEAR) Ontology, AOP Ontology, and others can be found by searching the following ontology portals: | ||||||||||||||||
Semantics | The meaning of a string (e.g., words, phrases, sentences) in a language; of or relating to the study of meaning and changes of meaning. [Source: NCIt C54194] | Learn how NCI is using semantics to build interoperable systems accessible to both humans and machines. | ||||||||||||||||
Syntax | The rules (word order, punctuation, sentence structure, etc.) for writing a language. As applied in computer science, it refers to the structure needed for a computer to read and understand the coded instructions or information to perform a task. [Source: Wikipedia] | Languages for programming (Java, Python, …), mark-up (HMTL, JSON, …), and knowledge representation (OWL, RDF, …) each have their own syntax for coding. | ||||||||||||||||
Taxonomy | A taxonomy (or taxonomical classification) is a scheme of classification, with a tree-based hierarchical structure showing the relationships (parent/child or broader/narrow) of terms with each other within the taxonomy. Taxonomies typically lack the more complex relationships found in thesauri or ontologies. [Source: About Taxonomies & Controlled Vocabularies] | Integrated Taxonomic Information System is based on the Linnaean taxonomy for classification of organisms. Other biomedical examples include the International Classification of Disease and NCBI Taxonomy. | ||||||||||||||||
Thesaurus | A thesaurus is an extension of a taxonomy. At its base is a standard hierarchical structure showing broader/narrower term relationships. In addition, a thesaurus also shows associative (see also), and equivalent (use/used from or see/seen from) term relationships. It is common in thesauri that some or all terms have scope notes, which are brief explanations of how the term should be used. [Source: About Taxonomies & Controlled Vocabularies] | Examples of thesauri include NCBI’s Medical Subject Headings (MeSH) and NCI Thesaurus. |
 Common Language Glossary
Environmental Health Language Collaborative
Common Language Glossary
Environmental Health Language Collaborative
to Top
Last Reviewed: February 17, 2026


