DNA Repair X-Ray Crystallography

Robert S. Williams, Ph.D.
[email protected]
Research Summary
Scott Williams, Ph.D., is Deputy Chief of the Genome Integrity and Structural Biology Laboratory, head of the Structural Cell Biology Group, and holds a secondary appointment in the NIEHS Signal Transduction Laboratory.
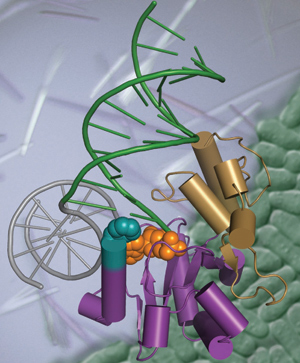
Exposure to environmental toxicants and stressors, pharmaceutical drugs, chronic inflammation, cellular respiration, and routine DNA metabolism all contribute to the production of cytotoxic DNA strand breaks. We focus on understanding the mechanisms through which chemically heterogeneous DNA breaks are recognized and repaired in cells, and elucidating how DNA repair complexes integrate with the cellular signaling apparatus to signal DNA damage. The group utilizes a multidisciplinary approach by combining high-resolution (X-ray crystallography) and low-resolution (Small angle X-ray scattering) macromolecular structural methods with biochemical and genetic studies to understand:
- How DNA damage is recognized, repaired and signaled, using atomic resolution.
- How proteins that guard stability of the genome are impacted by mutation in cancer predisposition syndromes and neurological diseases.
- How the cellular DNA repair machinery can be targeted for the improved treatment of human diseases including cancer.
Major areas of research:
- Structure and function of proteins that recognize, repair and signal DNA single and double strand breaks.
- Post-translationally regulated (e.g. phosphorylation, Adp-ribosylation) protein assembly.
- Mechanisms of DNA repair factor inactivation in heritable diseases.
Recent accomplishments:
- Uncovered molecular bases for DNA damage recognition by Aprataxin (Aptx) that defined a mechanism of the Aptx deadenylase in protecting genome integrity, and testable hypotheses for how Aptx is inactivated in the heritable neurodegenerative disorder Ataxia with Oculomotor Apraxia.
- Dissected structures and mechanisms for members of the multi-protein DNA double strand break sensing and processing Mre11/Rad50/Nbs1/CtIP complex that provide insights into Mre11 and Nbs1 inactivation in Ataxia Telangiectasia like (ATLD) disorder, and the radiation sensitivity disease Nijmegen breakage syndrome (NBS).
Williams earned his Ph.D. in biochemistry in 2003 from the University of Alberta, Canada, and completed his postdoctoral training at the Scripps Research Institute in La Jolla, Calif. before joining NIEHS in November 2009. Williams was named Deputy Chief of the Genome Integrity and Structural Biology Laboratory in 2016.
Contact
- Lab phone: (919) 316-4682


