1. Introduction
We implemented 3 algorithms for tag SNP selection in TAGster. These algorithms are:
Algorithm 1: A Greedy algorithm for single or multi-population tag SNP;
Algorithm 2: An efficient exhaustive search algorithm for single population tag SNP;
Algorithm 3: A two-stage solution algorithm for multi-population tag SNP.
We evaluated these algorithms against algorithms in existing software ldSelect (Carlson et al. 2004), FESTA (Qin, et al. 2006) and MultiPop-TagSelect (Howie, et al. 2006) using SNP genotype data from Environmental Genome Project(EGP).
2. Data
2.1 EGP Panel 2
At the time of this study, 207 genes were resequenced by EGP across 95 DNA samples from 4 populations (27 Africans, 24 Asians, 22 Europeans, and 22 Hispanics). There were a total of 16,153 SNPs with minor allele frequency (MAF) ≥ 0.05 in at least one of the 4 populations.
2.2 HapMap ENCODE
HapMap ENCODE (Encyclopedia of DNA Elements) Project resequenced ten 500 kb genomic regions in 48 individuals and subsequently genotyped all discovered SNPs as well as all SNPs in dbSNP at the time in 270 HapMap DNA samples from 3 populations including 30 CEPH (Utah residents with ancestry from northern and western Europe) trios, 90 Asians (45 unrelated JPT (Japanese in Tokyo, Japan), 45 unrelated CHB (Han Chinese in Beijing, China) and 30 YRI (Yoruba from Ibadan, Nigeria) trios. There were a total of 11,700 SNPs with minor allele frequency (MAF) ≥ 0.05 in at least one of the 3 populations.
3. Single Population Tag SNP
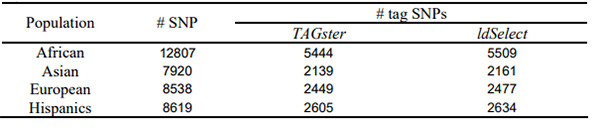
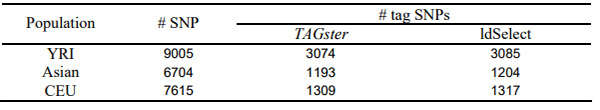
We applied both the refined greedy algorithm in TAGster and the greedy algorithm in ldSelect to select population specific tag SNPs at r2 threshold of 0.8 from each population specific data set. Table 1 shows that, in EGP data, the modified greedy algorithm selected l42 fewer tag SNPs than the greedy algorithm as implemented in ldSelect (Carlson, et al., 2004) in EGP. For 62 genes the modified greedy algorithm selected fewer tags in at least one of the 4 populations, whereas the greedy algorithm had fewer tag SNPs in only 2 genes in one population. Table 2 shows the modified greedy algorithm selected 30 fewer tag SNPs than ldSelect using HapMap ENCODE data.
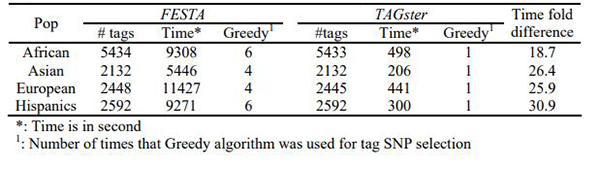
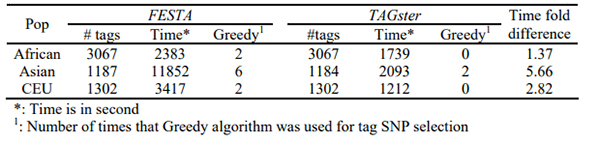
We applied both the exhaustive search algorithms in TAGster and the comprehensive search algorithm in FESTA (Qin, et al., 2006) to select population specific tag SNPs at r2 threshold of 0.8 and an exhaustive search step limit specification of 1,000,000 (the default setup of FESTA) for both algorithms for each of the 4 populations in EGP Panel 2.
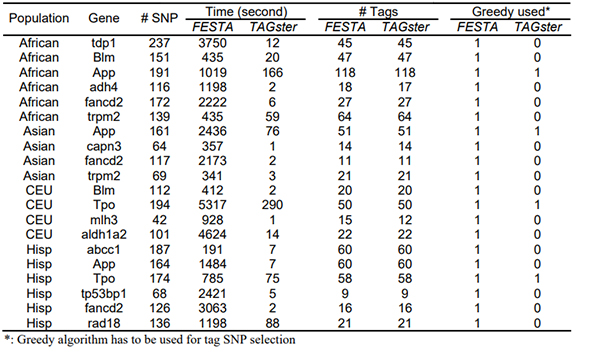
Table 3 shows that the exhaustive search algorithm in TAGster greatly improved the computational efficiency in all 4 populations. Moreover, FESTA did not find an optimal solution for the number of tag SNPs for 1 gene in Africans and 1 gene in Europeans. FESTA exceeded the 1,000,000 step limit and defaulted to use of the greedy algorithm 20 times in order to provide a result while TAGster only used greedy algorithm 4 times (Table 4). Evaluation of HapMap ENCODE data to generate table 5 showed a similar pattern of computational efficiency and requirements for defaulting to the greedy algorithm.
4. Multiple Population Tag SNP
We applied the modified greedy algorithm (Algorithm 1) and 2-stage method (Algorithm 3) to select multi-population tag SNP in 207 genes for the 4 populations from EGP Panel 2 and used as a benchmark measure the number of tag SNPs found using ldSelect followed by MultiPop-TagSelect (Howie, et al., 2006). The generalized modified greedy algorithm (generalized algorithm 1 for multiple populations) reduced tag SNP requirements by 183 SNPs whereas the two-stage method (Algorithm 3) reduced tag SNP requirements by 159 SNPs. If for each gene we selected the minimum of these two methods, it reduced tag SNP requirements by 233 SNPs below that required by ldSelect followed by MultiPop-TagSelect (Table 4). Evaluation in 3 populations from HapMap ENCODE shows a similar pattern of reduction (Table 6)
Both TAGster and MultiPop-TagSelect allow an investigator to specify a priori a set of SNPs for inclusion as tag SNP. MultiPop-TagSelect algorithm selects from population specific tag SNPs. Thus if an investigator-specified SNP is not one of these population specific tag SNPs, then it can not serve as a proxy for any population specific LD bin. Conversely, in the TAGster selection process, every investigator-specified SNP can serve as a proxy for other SNPs unless it is a singleton SNPs.


5. Multiple SNP Bin Tag SNP
In order to further reduce the number of tag SNPs, investigators may choose to select tag SNPs only for bins that contain multiple SNPs. The minimum bin size can be specify using the parameter -minimum in the parameter file params.txt. For example setting -minimum: 2 requires that bins contain at least two SNPs and eliminates singleton bin tag SNPs. Elimination of singleton bin tag SNPs can dramatically cut down the number of tag SNPs, while still capturing the majority of SNPs. It is particularly useful when selecting multiple population tag SNPs. For example, if parameter –minimum is set to a value of 2, TAGster selected 4094 multiple population multiple SNP bin (MPMS) tag SNPs for the 4 populations in EGP, compared to 7429 SNPs required if singleton bins are tagged. This smaller number of tag SNPs still captures ~95% common SNPs in Asian and CEPH populations, 91% in Hispanic population and 84% in Africans. For HapMap ENCODE data, 2095 MPMS tag SNPs (out of total of 3882 tag SNPs if singleton bin tags are included) can capture ~96% of common SNPs in Asian and CEU and 86% of SNPs in YRI.


