Features
Precision Environmental Health
Precision Environmental Health seeks to prevent disease by understanding risks and tailoring interventions to individuals. SRP researchers are at the forefront of this work and well-equipped to advance it. This issue showcases some SRP research projects that embody a precision environmental health approach. These projects leverage data science to link diverse data types, investigate how exposures affect health, and explore why susceptibility varies in people.
Understanding the Basis of Interindividual Differences in Susceptibility
Researchers at the Texas A&M University (TAMU) SRP Center are developing a translational in vitro-to-in vivo strategy to quantitatively predict and characterize the health hazards of complex environmental exposures. Led by Ivan Rusyn, Ph.D., their approach combines lab-based, or in-vitro, studies with population-based exposure data to understand how complex environmental exposures affect tissues and people differently. The team tested their approach on human cells derived from various populations and found it has potential to fill gaps in risk assessment data. The TAMU SRP Center used this method to evaluate how PFAS-mixtures may impact heart muscle cells and how contaminant mixtures vary in how toxic they are to blood cells from diverse subpopulations. Another TAMU study showed that incorporating in vitro data improved the precision of reference doses used for chemical risk assessments.
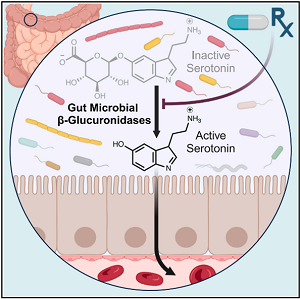
This figure from Lu's research team demonstrates how the gut microbiome influences hormone balance, and how prescription drugs can interfere with that process. (Image adapted from Simpson et al., 2024)
Led by Kun Lu, Ph.D., scientists at the University of North Carolina at Chapel Hill (UNC) SRP Center are using a precision environmental health approach to understand how the gut microbiome influences inorganic arsenic-induced diabetes. Their studies in mice have revealed individual differences in susceptibility, such as changes in body composition interacting with arsenic exposure to increase biological markers of type 2 diabetes markers. Using multi-omic, in vitro, and animal studies, the team also found that gut microbial enzymes play a crucial role in maintaining hormone balance, influencing individual differences in disease onset and treatment.
University of Oregon SRP Center scientists are developing an approach to measure how differences in tissue composition, body composition, and the properties of polycyclic aromatic hydrocarbons (PAH) affect human’s susceptibility to PAHs. Led by Jordan Smith, Ph.D., of the Pacific Northwest National Laboratory, the team developed a framework for analyzing rare non-coding genes linked to complex human diseases and identified key predictors for personal chemical exposures. They also used mass-spectrometry techniques to illustrate how dietary interventions can alter carcinogens’ toxicity.
Louisiana State University SRP Center researchers developed a stress response model that helped outline how respiratory system cells respond to oxidative stress. They found that healthy cells are significantly less susceptible to oxidative stress than unhealthy cells, and they contribute these differences to oxidative stress activating different pathways.
Developing Biomarkers of Exposure and Disease Risk
Researchers at the Harvard School of Public Health SRP Center are studying how prenatal and postnatal metal exposures affect cognitive health later in life. Using data from a Colorado birth cohort, they modeled the relationship between prenatal particulate matter (PM2.5) exposure and birth weight. They found that higher prenatal PM2.5 exposure was linked to lower birth weight, with a stronger association among children of non-Hispanic mothers with low educational status, younger age, or higher body mass index.
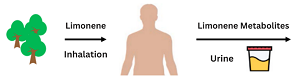
A visual representation of limonene inhalation leading to limonene metabolites in urine. (Adapted from Xie et al., 2024)
University of Louisville SRP Center scientists are using metabolites to study exposure to greenery on an individual level. They determined that limonene, an organic compound emitted into the air by plants, could be used as a biomarker of exposure to greenness. Researchers suspected that exposure to greenness could be measured by limonene metabolites in urine. They analyzed urine samples before and after limonene exposure from eight study participants who inhaled limonene in a laboratory setting, and eight participants who were exposed to greenness in a forest to compare limonene metabolites between known exposure and suspected exposure. The urine samples of the participants exposed to greenness in a forest had an increase in 14 out of 18 limonene metabolites tested, indicating that those metabolites could be used to examine greenness exposure.
A team at the Columbia University SRP Center is studying the effects of DNA methylation, a DNA modification that influences gene expression, on health outcomes such as cancer and diabetes. Notably, they found that DNA methylation is associated with risk of developing blood cancers. The team hopes DNA methylation could serve as a screening tool for early cancer detection. In two recent studies, the Columbia investigators used data from the Strong Heart Study cohort to explore connections between insulin resistance and type-2 diabetes among American Indian populations. One study suggests that maternal arsenic exposure may increase the risk of type-2 diabetes in offspring later in life. Additionally, the team found that DNA methylation could provide insights into insulin activity, potentially offering an early marker for type-2 diabetes risk.
Columbia researchers are also using DNA methylation to determine the effect of contaminants on a person’s biological age and its impacts on aging-related diseases. For example, in a study that compared biological age to chronological age in Native American communities living in an area with metals contamination, they found that exposure to certain metals was linked to accelerated aging and may increase the risk of age-related diseases. In particular, the nonessential metals arsenic, cadmium, and tungsten, were linked to faster aging, while essential metals like selenium, zinc, and molybdenum correlated with slower aging. According to the researchers, these findings suggest that reducing exposure to toxic metals may help prevent age-related diseases and premature death in these.

Amanda Armijo, Ph.D., conducting lab work at the MIT SRP Center. (Photo courtesy of Amanda Armijo)
Researchers from the Massachusetts Institute of Technology (MIT) SRP Center discovered that mice exposed to nitrosodimethylamine (NDMA) had specific changes to their DNA. Led by 2022 Wetterhahn Award Winner Amanda Armijo, Ph.D., the team revealed that NDMA caused a “sawtooth” mutation pattern in DNA from liver and lung cells. According to Armijo, scientists could use this pattern to identify if someone had been exposed to NDMA before the person experiences adverse health effects.
Exploring the Epigenome to Advance Risk Prediction
University of California (UC), Berkeley SRP Center researchers are studying how exposure to certain chemicals disrupts the DNA methylation process, thus causing disease. Through examining deviations in methylation across all 23 gene pairs, an approach known as epigenome-wide study, the scientists can identify where methylation changes occur and their potential health effects.

Martyn Smith, Ph.D., director of the UC Berkely SRP Center, explained the links between benzene and luekemia during his 2024 Herbert E. Stokinger Award Presentation this past October.
One team is tracking how exposure to benzene and formaldehyde in the workplace disrupts blood cell DNA methylation, which the researchers hypothesize may explain these chemical’s link to certain leukemias. Using data from factory workers in China, they found that benzene and formaldehyde exposure altered DNA methylation across the genes. The team also isolated irregularities in a tumor-suppressor gene as a potential biomarker of exposure to formaldehyde.
Another group conducted an epigenome-wide association study to explore if DNA methylation mediates the protective effect of folate (vitamin B9) intake during pregnancy against childhood acute lymphoblastic leukemia. They analyzed blood samples and maternal diet information in the pre-pregnancy and early pregnancy period, collected during a 13-years-long California study, and found that total maternal folate intake appears to reduce lymphoblastic leukemia risk across all populations. Further, they discovered that Hispanic participants with low income and educational levels showed a sharper increase in benefits from higher folate intake than other groups, indicating a potential benefit to targeting these groups for folate supplementation efforts.
UC Berkeley SRP researchers also looked at DNA methylation changes linked to arsenic exposure from drinking water, finding that methylation disfunction may underlie arsenic’s toxic effects. The team analyzed DNA methylation data from arsenic-exposed populations in Chile and Bangladesh. Through a two-step method of data harmonizing and processing combined with meta-analysis, they identified multiple locations on various genes where faulty methylation occurred after arsenic exposure. According to the researchers, their findings suggest DNA methylation is likely the mechanism controlling arsenic’s toxicity to humans and the patterns they found could serve as biomarkers for past exposure and to predict future disease risk.
to Top



