Overview
Expression Predictor (ExP) is a stand-alone, desktop application developed with Java™ programming language for classifying and predicting samples based on gene expression data using a simplified fuzzy adaptive resonance theory map (SFAM) neural network architecture (Kasuba, 1993). Leave-one-out cross validation of the samples at successive vigilance parameter settings permits the determination of the classifier setting that gives the highest accuracy of prediction. Gene expression data can be imported into ExP using a standard format for analysis and the prediction results exported in a tab-delimited format.
Specifics of the Methodology
Details of the Fuzzy ARTMAP algorithm and architecture have been made previously available (Azuaje, 2001; Downs et al., 1996; Carpenter et al., 1992) and the method has been optimized for simplification (SFAM; Kasuba, 1993). Normalization of the ith input gene expression pattern vector, ai, of d gene features was achieved and represented by computation of the complement-coded input vector I such that
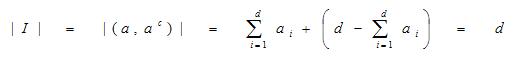 (1)
(1)
where aci = 1-ai . Complementation expands the input to twice its size. Following input normalization , activation of the jth output node (encodes a category or class of samples) based on its weights wj and choice parameter alpha, ?, set very low (< 0.01) is denoted as Tj where
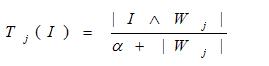 (2)
(2)
and the winning output node, Winner = max (Tj). The minimum symbol ? is referred to as the 'and' operator in fuzzy logic. Since | I | is equal to d in equation (1), the match function to compare complement-coded input features and an output node's weight is
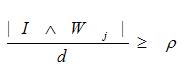 (3)
(3)
where ?? is the vigilance parameter indicating the network encoding granularity (refinement). If the match function is greater than ? the network is in a state of resonance and the output node j is sufficient to encode input I. Finally, the weight vector Wj from the output node is updated with a learning rate ? set to 1.0 such that
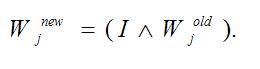 (4)
(4)
Feedback of the category nodes to the match tracking component permits supervised classification of the samples.
Requirements for ExP
The gene expression data needs to be normalized and transformed to the user’s satisfaction. In addition, missing data needs to be imputed. The first column of the data matrix needs to contain the gene IDs and the other columns the gene expression data. The first row of the data matrix needs to contain the experiment name and the second row needs to contain the text or numeric label representing the class of the sample. Finally, the software has been tested to run on Windows PCs running the XP operating system and requires JRE version 1.4.2 or later.
Downloads
- Download the ExP archive (74KB)
- View the Quick Start Guide
- Report bugs, corrections and suggestions to [email protected].
References
Azuaje, F. (2001) A computational neural approach to support the discovery of gene function and classes of cancer. IEEE Trans Biomed Eng., 48, 332-339.
Carpenter, G., Grossberg, S., Markuzon, N., Reynolds, J.H., and Rosen, D.B. (1992) Fuzzy artmap: a neural network architecture for incremental supervised learning of analog multidimensional maps. IEEE Transactions on Neural Networks, 3, 698-713.
Kasuba, T. (1993) Simplified fuzzy artmap. AI Expert, 8, 18-25.
Public Domain Notice
This is U.S. government work. Under 17 U.S.C. 105 no copyright is claimed and it may be freely distributed and copied.
Contact
-
Robert P. Bushel, Ph.D.
Special Volunteer -
Tel 919-618-1945
[email protected]


