Gene-Environment Interaction in Disease and Aging

Research Summary
Xiaoling Li, Ph.D., heads the Metabolism, Genes, and Environment Group and holds a secondary appointment in the NIEHS Epigenetics and RNA Biology Laboratory. The long-term goal of Metabolism, Genes, and Environment Group is to understand how cellular metabolism mediates environmental influences on cellular functions through interaction with epigenetic modification enzymes, and to further investigate how dysregulation of this interplay contributes to pathogenesis of diseases and aging.
To achieve this goal, the group focuses on a cellular metabolic sensor, SIRT1, and protein acetylation/acylation. SIRT1 belongs to a family of highly conserved NAD+-dependent protein modification enzymes called sirtuins. They are capable of removing a wide range of lipid lysine acyl-groups, including acetyl-group, from protein substrates in a NAD+-dependent manner. These NAD+-dependent activities enable sirtuins to monitor cellular energy status and modulate gene transcription, energy metabolism, and genome stability in response to environmental signals. These activities are also important for cell survival in response to various environmental stressors and are required for lifespan extension provided by calorie restriction (CR) in a number of model organisms. Therefore, sirtuins are essential genetic factors that directly link the environment to animal physiology, providing us a unique opportunity to study gene-environment interactions during the processes of disease and aging.
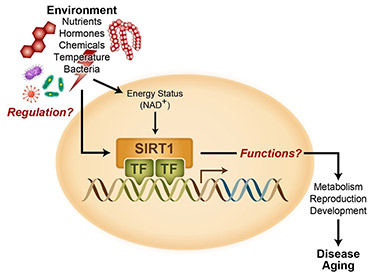
SIRT1, the most conserved mammalian member of sirtuin family, is a NAD+-dependent protein deacetylase/deacylase that target histones, transcription factors, splicing factors, as well as numerous transcription co-factors. Utilizing mice and culture cells as model systems, the group combines molecular, cellular, and genetic approaches to study the interplay of SIRT1, protein acetylation/acylation, and cellular metabolism in mediating environmental influence on aging, development, and human diseases. Recent studies from the group reveal that SIRT1 is a critical transcriptional regulator of intestinal nutrient metabolism (Kazgan et al., Gastroenterology, 2014), gut microbiota homeostasis and inflammation (Wellman et al., Gastroenterology, 2017), and intestinal tumorigenesis (Ren et al., Current Biology, 2017). The group further uncover that gut commensal bacteria cooperate with host cells to promote host NAD+ biosynthesis in response to dietary NAD+-boosting supplements (Shats et al., Cell Metabolism, 2020). Additionally, SIRT1, metabolism, and histone acylation are pivotal in regulation of embryonic stem cell pluripotency, differentiation, and animal development (Tang et al., Molecular Cell, 2014; Tang et al., EMBO J., 2017; Fang et al., Cell Stem Cell, 2020).
Major areas of research:
- Host-microbe interaction in host NAD+ metabolism and related (patho)physiology
- Host regulation of gut microbiota in intestinal tissue homeostasis, inflammation, and tumorigenesis
- Metabolic and epigenetic regulation of tumorigenesis
- Metabolic and epigenetic regulation of stem cell functions and embryogenesis
Current projects:
- Intestinal SIRT1 in regulation of gut microbiota and tissue homeostasis
- Regulation of host NAD+ metabolism by manipulating gut microbiome
- SIRT1 and methionine metabolism in stem cell maintenance and embryogenesis
- SIRT1, metabolism, and protein acylation in regulation of stem cell differentiation
- Dietary methionine restriction, cellular methionine metabolism and their impact on redox homeostasis, chromatin, and tumor biology
Li received her Ph.D. in Biological Chemistry from the Johns Hopkins School of Medicine in 2002. She was a Leukemia & Lymphoma Society postdoctoral fellow in the laboratory of Leonard Guarente at Massachusetts Institute of Technology before joining NIEHS in 2007.


