Nuclear Protein Physiology

Anton M. Jetten, Ph.D.
[email protected]
Research Summary
Anton M. Jetten, Ph.D., is Head of the Cell Biology Group in the Immunity, Inflammation, and Disease Laboratory, and holds a secondary appointment in the NIEHS Epigenetics and RNA Biology Laboratory. Research carried out by members of the Cell Biology Group has led to the discovery of several novel transcriptional regulators: the GLI-Similar 1-3 (GLIS1-3) Krüppel-like zinc finger transcription factors (Project I) and the retinoic acid-related orphan nuclear receptor γ (RORγ; Project II). These proteins play a critical role in the regulation of many physiological processes and are implicated in several environmentally relevant pathologies (e.g., diabetes, cancer, metabolic syndrome, neurological disorders, inflammatory diseases). The goal of this research is to elucidate the molecular mechanisms by which these proteins regulate gene transcription to obtain insights into how they control various biological processes and are implicated in disease. These studies might lead to novel therapeutic strategies in the management of several diseases.
Project I: GLIS1-3 Transcription Factors: Study of their Transcriptional Activity, Physiological Functions, and Roles in Disease
The GLI-Similar 1-3 (GLIS1-3) proteins form a subfamily of Krüppel-like zinc finger transcription factors discovered by our laboratory. They can function as activators or repressors of gene transcription by interacting with G-rich GLIS binding sites (GLISBS) in the promoter of target genes. GLIS2/3 transcriptional activity appears to be controlled by primary cilium-associated signaling pathways. GLIS proteins regulate gene transcription in coordination with other transcription factors. GLIS3 plays a critical role in the regulation of many biological processes and several diseases that are major global health concerns. We demonstrated that GLIS3 is essential for the generation of pancreatic beta cells and insulin gene transcription, thyroid hormone biosynthesis, early spermatogenesis, and the maintenance of normal kidney functions. GLIS3 promotes differentiation of human pluripotent stem cells (hPSCs) into posterior neural progenitor cells by activating WNT3A transcription. In humans and mice, GLIS3 deficiency causes neonatal diabetes, congenital hypothyroidism, glaucoma, inflammation, fibrosis, neurological disorders, male infertility, and polycystic cystic kidney disease. Loss of GLIS2 function causes nephronophthisis, a cystic renal disease, that is characterized by renal atrophy, fibrosis, and inflammation. GLIS2 regulates stem cell renewal and is implicated in metabolic syndrome, leukemia, and breast cancer. GLIS1 is a critical regulator of intraocular pressure and has been implicated in glaucoma and cancer. To obtain further insights into their regulation of gene transcription and biological functions of GLIS proteins, our current research is focusing on identifying the role of GLIS1-3 in the regulation of cell-type specific functions and chromatin structure using single cell sequencing and ATAC-Seq, and cell type-specific knockout and overexpression of GLIS1-3 in mice. Our studies suggest that GLIS1-3 pathways might provide novel therapeutic targets in the management of multiple diseases.
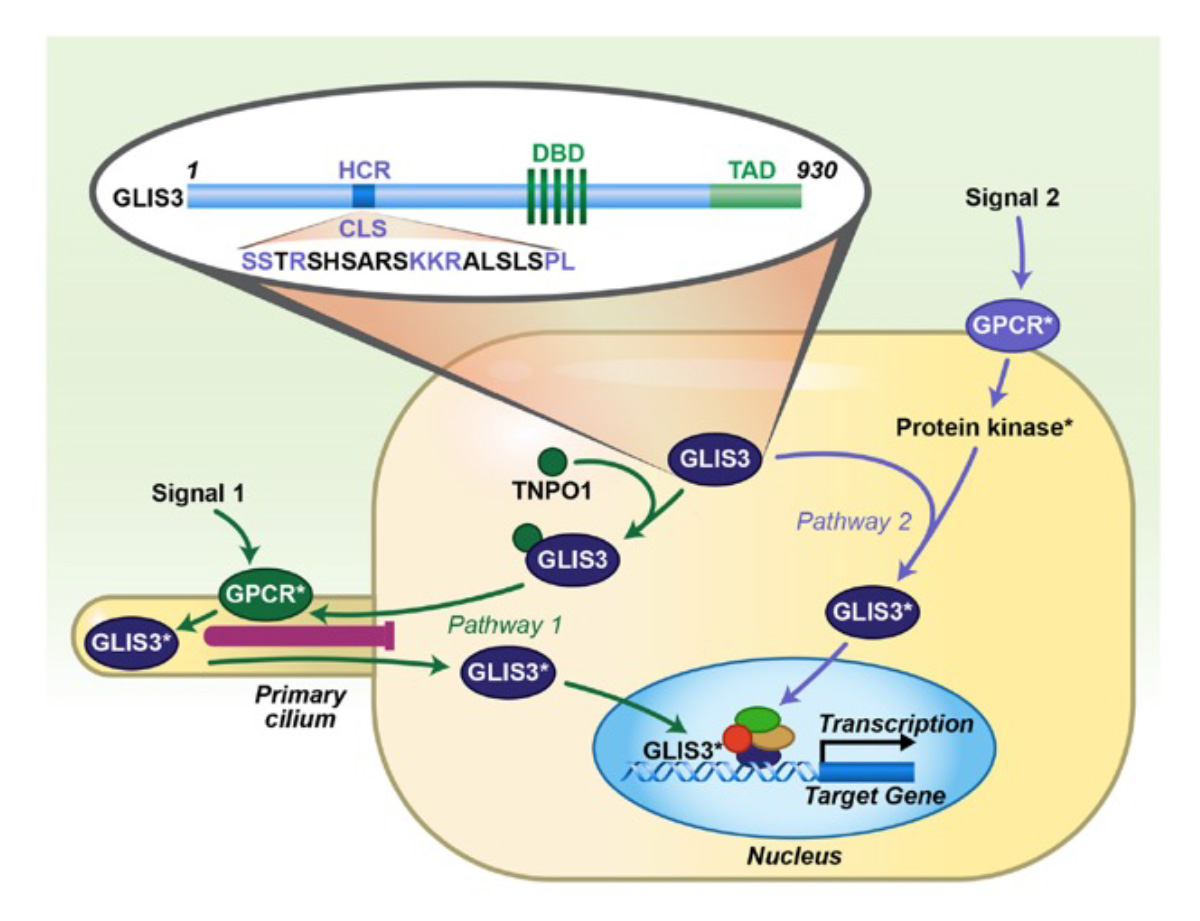
Project II: ROR Nuclear Receptor Signaling: Mechanism of Action, Physiological Functions, Roles in Disease, and Potential Therapy.
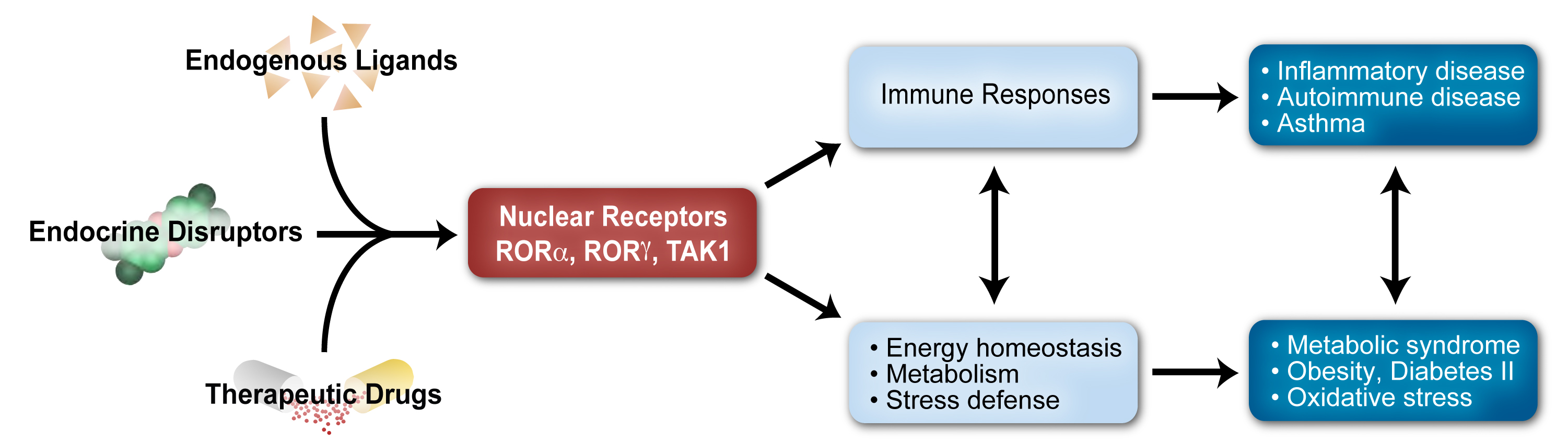
The research in this project is focusing on the study of the retinoic acid-related orphan receptors RORα and RORγ, the latter of which was discovered by our laboratory. RORs regulate transcription by binding as monomers to ROR response elements (ROREs) consisting of the RGGTCA consensus, in the regulatory regions of target genes. RORs act as ligand-dependent transcription factors. A series of sterol metabolites bind RORs and function as agonists or inverse agonists modulating either positively or negatively ROR transcriptional activity and consequently physiological processes regulated by RORs. RORs play a critical role in the regulation of many physiological processes, including embryonic development, immunity, various metabolic pathways, and neural processes, and are relevant to an increasing number of pathologies, including autoimmune disease, metabolic syndrome, cancer, and various neurological disorders. Thus, RORγ inverse agonists might be useful in the management of several inflammatory and endocrine disorders.
Major areas of research:
- Identification of the molecular mechanisms by which GLIS1-3 regulate gene transcription and how this relates to their function in several tissues (pancreas, kidney, thyroid, lung, brain).
- Identification of (primary cilium-associated) upstream pathways that regulate GLIS1-3 transcriptional activity through various posttranslational modifications (e.g., phosphorylation).
- Obtain further insights into the roles of GLIS1-3 in cancer, and several inflammatory and endocrine disorders.
- Determine the role of GLIS proteins in the regulation of differentiation and proliferation in stem and progenitor cells.
- Identifying the molecular mechanisms of the role of RORs in endocrine and inflammatory disease.
Current projects:
- Study the role of GLIS2/3 in the regulation of gene transcription in the kidney and lung in relation to cystic kidney disease and lung fibrosis and inflammation.
- Study the role of GLIS3 in the regulation of gene transcription in the thyroid gland in relation to its role in hypothyroidism.
- Study of the role of GLIS3 and GLIS2 in pancreatic islets and ductal cells using scRNA-Seq and ChIP-Seq, and various knockout and knockin mouse models.
- Characterize the role of GLIS3 in the differentiation of neural stem cells along different lineages.
- Identifying the molecular mechanisms of the role of GLIS proteins in cancer.
- Identification of the functions of RORα and RORγ in the pancreas and neural cells in relation to their role in endocrine and neurological disorders.
Relevance to NIEHS Mission
Members of the GLIS and ROR subfamilies of transcription factors are implicated in several environmentally relevant diseases (e.g., cancer, diabetes, metabolic syndrome, glaucoma, autoimmune disease) in which environmental factors (e.g., high fat/carbohydrate diet, UV light, endocrine disruptors, pathogens) are important risk factors. The research program of the Cell Biology Group uses in vitro cell and in vivo mouse models as well as human epidemiological studies, to obtain greater insights into the causal mechanisms and progression of these pathologies, the role of environmental factors in them, and examine the potential of these proteins as novel therapeutic targets in the management of these diseases.


