Transcriptional & Epigenetic Control of Cell Fate Decisions

Research Summary
Raja Jothi, Ph.D., heads the Systems Biology Group within the Epigenetics and RNA Biology Laboratory, and holds a secondary appointment in the NIEHS Biostatistics and Computational Biology Branch.
Understanding how environmental cues affect cell physiology to give rise to specific phenotypic outcomes is a fundamental step toward understanding disease mechanisms. During development and in response to environmental insults, various signaling cascades culminate in the activation of appropriate master transcription factors (TFs) and chromatin remodeling enzymes to establish gene expression programs controlling cell fate decisions. Elucidation of gene networks (Figure 1) associated with specific biological phenotypes thus becomes key to understanding the molecular basis of development and pathogenesis.
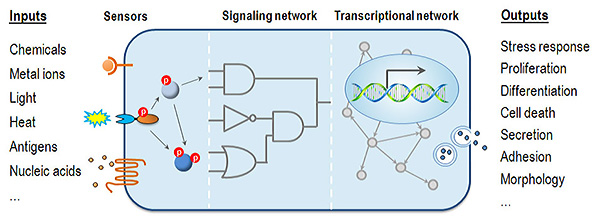
The long-term goal of the Jothi Group is to understand the fundamental principles of transcriptional and epigenetic control of cell fate decisions during development and pathogenesis and to apply this knowledge to aid the development of diagnostic and therapeutic strategies for cancer and other diseases.
Over the past decade or so, the group’s focus has been on understanding how signaling dynamics instruct epigenetic and/or transcriptional programs controlling cell fate decisions during pre- to post-implantation embryo development. These investigations use mouse embryonic stem cells (ESCs) and defined culture conditions for in vitro ESC differentiation to model early embryo development. ESCs, derived from the inner cell mass of the blastocyst, are an ideal model system for understanding gene networks controlling cell fate transitions. Their ability to self-renew indefinitely and differentiate into all derivatives of the three germ layers makes them an attractive model for exposure studies, disease modeling, and regenerative medicine. Notably, differentiated derivatives of human pluripotent cells could serve as excellent models for understanding disease mechanisms, particularly when there is no good animal model for the disease being studied.
The group uses integrative interdisciplinary approaches―merging computational biology, functional genomics, proteomics, and molecular biology―to map, reconstruct, and characterize developmentally and environmentally responsive gene regulatory networks and signaling pathways that underlie cellular identity and response. Over the years, the group has uncovered and characterized many genes and mechanisms with previously unknown roles in regulating cell fate decisions during development and tumorigenesis.
Major areas of research:
- Chromatin, epigenetics and gene regulatory networks
- Cell fate decisions during development and tumorigenesis
- Methods development for integration and analysis of Big Data
Current projects:
- Reconstruction and characterization of gene networks regulating stem cell identity
- Investigation into how signaling cascades instruct epigenetic or transcriptional programs controlling cell fate
- Intragenic enhancers and transcription regulation
Jothi received his Bachelor’s degree in Computer Science & Engineering from the University of Madras in 1998, and his Ph.D. in Computer Science & Discrete Mathematics from the University of Texas at Dallas in 2004. Following postdoctoral training in Computational Biology and Epigenetics at the National Center for Biotechnology Information (NCBI, NIH) and the National Heart, Lung, and Blood Institute (NHLBI, NIH), respectively, he was recruited to NIEHS in 2009 and promoted to Senior Investigator with tenure in 2015. Dr. Jothi was the recipient of the NIEHS Early Career “Rising Star” Award in 2009 and the NIH Ruth L. Kirschstein Mentoring Award in 2016.


