Introduction

For decades, scientists have known the basic structure of our DNA, the building blocks that make up our genes. Although nearly every cell in the human body has the same set of genes, why is it that different types of cells, such as those from brain or skin, look and behave so differently?
The answer is epigenetics, a rapidly growing area of science that focuses on the processes that help direct when individual genes are turned on or off. While the cell’s DNA provides the instruction manual, genes also need specific instructions. In essence, epigenetic processes tell the cell to read specific pages of the instruction manual at distinct times.
Some epigenetic changes are stable and last a lifetime, and some may be passed on from one generation to the next, without changing the genes.
What is NIEHS Doing?

NIEHS is currently supporting epigenetics research that is accelerating the understanding of human biology and the role of the environment in disease. These discoveries may lead to the development of new ways to prevent and treat diseases for which the environment is believed to be a factor.
NIEHS at the forefront of epigenetics research
In 2003, NIEHS-supported researchers made an important discovery that demonstrated the role of environmental epigenetics in development and disease.2 They used the agouti mouse in their study. The mouse has an altered version of the agouti gene, which causes them to be yellow, obese, and highly susceptible to developing diseases, such as cancer and diabetes.
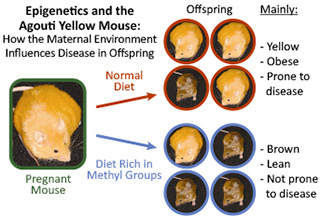
The researchers fed the mice a diet rich in methyl groups. Through epigenetic processes, the methyl groups attached to the mother’s DNA, and turned off the agouti gene. As a result, most of the offspring were born lean and brown, and no longer prone to disease.
This study was the first to demonstrate that it is not just our genes that determine our health, but also our environment and what we eat.
NIEHS-supported researchers establish epigenomic reference maps
While researchers have known, for quite some time, the sequence of DNA that make up all human genes, collectively known as the genome, the same could not be said for the human epigenome, until recently. The epigenome refers to all of the chemical compounds added to the genetic material of an organism that regulate its function.
NIEHS and the National Institute on Drug Abuse (NIDA) co-led a national effort, through the NIH Roadmap Epigenomics Program, to create a series of epigenomic maps representing locations on the DNA where chemical compounds attached in more than 100 different tissue and cell types, including blood, lung, heart, gastrointestinal tract, brain, and stem cells. The groundbreaking work was featured in a 2015 article in the journal Nature.3
By comparing the epigenomic map of a healthy cell or tissue, with the map of the same cell or tissue after an environmental exposure or in relation to a specific disease, NIEHS scientists can better understand how the environment affects genes through epigenetic processes. The epigenomic maps are available to the entire scientific community through the Washington University Epigenome Browser.
Grantees study link between environmental pollution and disease
NIEHS-supported researchers at Harvard T.H. Chan School of Public Health have shown that human exposure to environmental air pollutants and toxic metals, such as arsenic, can cause damage to cells that may lead to cardiovascular disease. The research team has been tracking abnormalities in blood, as well as epigenetic changes, which may serve as indicators, or markers, of exposure to air pollution and toxic metals at levels that can increase the risk of cardiovascular disease, particularly in elderly men. These markers may help in the early detection and prevention of cardiovascular and other diseases.4 ,5
Grantees at Beth Israel Deaconess Medical Center used epigenetics to define the link between environmental exposure to smoke, mercury, and lead, and reproductive outcomes, such as preterm birth.6 ,7 Their results are important, since more than one in 10 infants worldwide is born prematurely, increasing their chances of having health problems later in life.8 The study also found that epigenetic changes may serve as markers for maternal chemical exposure during pregnancy.
Scientists investigate epigenetic control of genes involved in cancer
NIEHS scientists in the Epigenetics and RNA Biology Laboratory are examining how epigenetic mechanisms influence normal cell development, and contribute to biological processes involved in breast cancer and immune function. To date, they have uncovered many details of how the protein complex Mi-2/NuRD controls genes involved in breast cancer. Mi-2/NuRD is found in the nucleus of cells and includes enzymes that affect histone modifications that regulate gene activity.
The research is helping to identify what genes are regulated by the complex and how the complex alters epigenetic processes that may contribute to disease. Understanding the underlying mechanisms of disease may lead to the development of new methods to diagnose, prevent, and treat diseases, such as breast cancer, in the future.
Researchers unravel role of epigenetics in development, inheritance, and disease
NIEHS-supported researchers have found that early-life exposure to nutritional and dietary factors, maternal stress, and environmental chemicals can increase the likelihood of developing disease and poor health outcomes later in life. In addition, some of the effects of these exposures can be passed down for multiple generations, even after the original exposure has been removed, through a process known as transgenerational inheritance.
Researchers in the NIEHS Transgenerational Inheritance in Mammals After Environmental Exposure (TIME) Program are using mice and rats to investigate how transgenerational inheritance occurs after exposure to environmental exposures, whether the process is different in males and females, and when in development these events are most likely to occur. Understanding how transgenerational inheritance of effects from exposures occur in animals may shed light on similar processes in humans.
-
Waterland RA, Jirtle RL. 2003. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol 23(15):5293-5300. [Abstract Waterland RA, Jirtle RL. 2003. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol 23(15):5293-5300.]
-
Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu YC, Pfenning AR, Wang X, Claussnitzer M, Liu Y, Coarfa C, Harris RA, Shoresh N, Epstein CB, Gjoneska E, Leung D, Xie W, Hawkins RD, Lister R, Hong C, Gascard P, Mungall AJ, Moore R, Chuah E, Tam A, Canfield TK, Hansen RS, Kaul R, Sabo PJ, Bansal MS, Carles A, Dixon JR, Farh KH, Feizi S, Karlic R, Kim AR, Kulkarni A, Li D, Lowdon R, Elliott G, Mercer TR, Neph SJ, Onuchic V, Polak P, Rajagopal N, Ray P, Sallari RC, Siebenthall KT, Sinnott-Armstrong NA, Stevens M, Thurman RE, Wu J, Zhang B, Zhou X, Beaudet AE, Boyer LA, De Jager PL, Farnham PJ, Fisher SJ, Haussler D, Jones SJ, Li W, Marra MA, McManus MT, Sunyaev S, Thomson JA, Tlsty TD, Tsai LH, Wang W, Waterland RA, Zhang MQ, Chadwick LH, Bernstein BE, Costello JF, Ecker JR, Hirst M, Meissner A, Milosavljevic A, Ren B, Stamatoyannopoulos JA, Wang T, Kellis M. Integrative analysis of 111 reference human epigenomes. Nature 518(7539):317-330. [Abstract Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu YC, Pfenning AR, Wang X, Claussnitzer M, Liu Y, Coarfa C, Harris RA, Shoresh N, Epstein CB, Gjoneska E, Leung D, Xie W, Hawkins RD, Lister R, Hong C, Gascard P, Mungall AJ, Moore R, Chuah E, Tam A, Canfield TK, Hansen RS, Kaul R, Sabo PJ, Bansal MS, Carles A, Dixon JR, Farh KH, Feizi S, Karlic R, Kim AR, Kulkarni A, Li D, Lowdon R, Elliott G, Mercer TR, Neph SJ, Onuchic V, Polak P, Rajagopal N, Ray P, Sallari RC, Siebenthall KT, Sinnott-Armstrong NA, Stevens M, Thurman RE, Wu J, Zhang B, Zhou X, Beaudet AE, Boyer LA, De Jager PL, Farnham PJ, Fisher SJ, Haussler D, Jones SJ, Li W, Marra MA, McManus MT, Sunyaev S, Thomson JA, Tlsty TD, Tsai LH, Wang W, Waterland RA, Zhang MQ, Chadwick LH, Bernstein BE, Costello JF, Ecker JR, Hirst M, Meissner A, Milosavljevic A, Ren B, Stamatoyannopoulos JA, Wang T, Kellis M. Integrative analysis of 111 reference human epigenomes. Nature 518(7539):317-330.]
-
Bind MA, Baccarelli A, Zanobetti A, Tarantini L, Suh H, Vokonas P, Schwartz J. 2012. Air pollution and markers of coagulation, inflammation, and endothelial function: associations and epigene-environment interactions in an elderly cohort. Epidemiology 23(2):332-340. [Abstract Bind MA, Baccarelli A, Zanobetti A, Tarantini L, Suh H, Vokonas P, Schwartz J. 2012. Air pollution and markers of coagulation, inflammation, and endothelial function: associations and epigene-environment interactions in an elderly cohort. Epidemiology 23(2):332-340.]
-
Bind MA, Lepeule J, Zanobetti A, Gasparrini A, Baccarelli A, Coull BA, Tarantini L, Vokonas PS, Koutrakis P, Schwartz J. 2014. Air pollution and gene-specific methylation in the Normative Aging Study: association, effect modification, and mediation analysis. Epigenetics 9(3):448-458. [Abstract Bind MA, Lepeule J, Zanobetti A, Gasparrini A, Baccarelli A, Coull BA, Tarantini L, Vokonas PS, Koutrakis P, Schwartz J. 2014. Air pollution and gene-specific methylation in the Normative Aging Study: association, effect modification, and mediation analysis. Epigenetics 9(3):448-458.]
-
Burris HH, Baccarelli AA, Motta V, Byun HM, Just AC, Mercado-Garcia A, Schwartz J, Svensson K, Téllez-Rojo MM, Wright RO. 2014. Association between length of gestation and cervical DNA methylation of PTGER2 and LINE 1-HS. Epigenetics 9(8):1083-1091. [Abstract Burris HH, Baccarelli AA, Motta V, Byun HM, Just AC, Mercado-Garcia A, Schwartz J, Svensson K, Téllez-Rojo MM, Wright RO. 2014. Association between length of gestation and cervical DNA methylation of PTGER2 and LINE 1-HS. Epigenetics 9(8):1083-1091.]
-
Sanders AP, Burris HH, Just AC, Motta V, Amarasiriwardena C, Svensson K, Oken E, Solano-Gonzalez M, Mercado-Garcia A, Pantic I, Schwartz J, Tellez-Rojo MM, Baccarelli AA, Wright RO. 2015. Altered miRNA expression in the cervix during pregnancy associated with lead and mercury exposure. Epigenomics 7(6):885-896. [Abstract Sanders AP, Burris HH, Just AC, Motta V, Amarasiriwardena C, Svensson K, Oken E, Solano-Gonzalez M, Mercado-Garcia A, Pantic I, Schwartz J, Tellez-Rojo MM, Baccarelli AA, Wright RO. 2015. Altered miRNA expression in the cervix during pregnancy associated with lead and mercury exposure. Epigenomics 7(6):885-896.]
-
Blencowe H, Cousens S, Oestergaard MZ, Chou D, Moller AB, Narwal R, Adler A, Vera Garcia C, Rohde S, Say L, Lawn JE, MRCP Paeds. 2012. National, regional, and worldwide estimates of preterm birth rates in the year 2010 with time trends since 1990 for selected countries: a systematic analysis and implications. Lancet 379(9832):2162-2172. [Abstract Blencowe H, Cousens S, Oestergaard MZ, Chou D, Moller AB, Narwal R, Adler A, Vera Garcia C, Rohde S, Say L, Lawn JE, MRCP Paeds. 2012. National, regional, and worldwide estimates of preterm birth rates in the year 2010 with time trends since 1990 for selected countries: a systematic analysis and implications. Lancet 379(9832):2162-2172.]
NIEHS Research Efforts
- Epigenetics and RNA Biology Lab
- Epigenetics Program
- Epigenetics Resources
- Toxicant Exposures and Response by Genomic and Epigenomic Regulators of Transcription (TaRGET) Program
- Transgenerational Inheritance in Mammals After Environmental Exposure Program (TIME) Program
Further Reading
Stories from the Environmental Factor (NIEHS Newsletter)
Additional Resources
- NIH National Human Genome Research Institute (NHGRI) Fact Sheet on Epigenomics
- Epigenetics Basics
- Epigenetics: How genes and the environment interact: In this one-hour videocast, Randy Jirtle, Ph.D., discusses the science of epigenetics as part of the NIH Director’s Wednesday Afternoon Lecture Series.


