Features
Bioremediation and the Microbiome - Expanding the Toolbox
Superfund Research Program (SRP) researchers study bacterial communities in the environment to understand how they can be used as a tool to clean up chemicals. They also study bacterial communities in our bodies to explore how exposure to potentially hazardous chemicals can alter the human microbiome and affect our health. Although the initial goals are different, some techniques and expertise can be similar. Both approaches improve our understanding of the mechanisms involved in how microbial communities interact with different contaminants.
Advancing Bioremediation
Bioremediation refers to the use of biota, including bacteria, algae, fungi, and plants, to reduce or detoxify hazardous substances in the environment. SRP researchers work to understand how microbes and plants break down contaminants into less toxic products and how factors in the environment can be altered to more efficiently break down hazardous substances.
Characterizing Microbial Communities
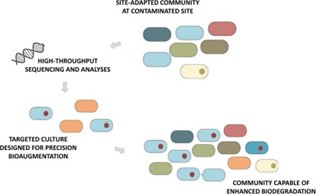
Duke University SRP Center researchers led by Claudia Gunsch, Ph.D., sequence the genomes of bacterial communities in soil and compare them to soil chemical profiles to identify bacteria that can survive in contaminated environments and enhance degradation of polycyclic aromatic hydrocarbons (PAHs). They recently characterized microbes that can break down creosote, a type of PAH commonly found at Superfund sites.
At the University of Iowa SRP Center, researchers are uncovering how plants break down polychlorinated biphenyls (PCBs) by identifying PCB-degrading microbes and the functional genes and enzymes involved. Led by Jerald Schnoor, Ph.D., they found PCB degradation hotspots in sediments from a contaminated wastewater lagoon in Virginia and identified specific genes associated with dechlorination.
Led by Lisa Alvarez Cohen, Ph.D., University of California, Berkeley SRP Center grantees evaluated how microbes from different classes or species work together to break down trichloroethene (TCE). They also study environmental conditions that facilitate TCE breakdown. They found differences in the composition and community dynamics of contaminant-degrading microorganisms with different levels of vitamin B12, indicating that different types of bacteria play a role in TCE degradation when B12 is limited.
SRP grantees study biological and geochemical conditions that can affect microbial communities and how well they can dechlorinate contaminants. Research led by Frank Loeffler, Ph.D., at the University of Tennessee showed that nitrous oxide ,a groundwater contaminant commonly generated from agricultural runoff, inhibits bacterial degradation of certain chlorinated contaminants, including tetrachloroethene (PCE). According to the researchers, these findings may explain why bioremediation can stall at some hazardous waste sites, particularly those with high levels of nitrous oxide.
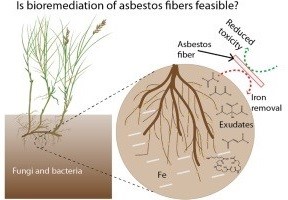
At the University of Pennsylvania (Penn) SRP Center, researchers discovered that natural compounds released from bacteria and fungi in soil, known as siderophores, can decrease the toxicity of asbestos fibers .According to the researchers, siderophores could potentially form the basis of a low-cost bioremediation strategy to manage asbestos-contaminated sites.
Understanding Mechanisms of Uptake and Bioavailability
At the University of New Mexico SRP Center, grantees explore how to improve the uptake of uranium and other metals into plants to immobilize the contaminants and prevent their release into community water sources. Led by Jose Cerrato, Ph.D., they recently found that calcium enables uranium transport into plant root cells.

In a field study, University of Arizona SRP Center researchers led by Raina Maier, Ph.D., showed that amending mine waste with compost in semi-arid conditions helped establish native plants and sustained their growth over 4 years. The approach, called compost-assisted phytostabilization, immobilizes the metals so they cannot be spread by wind or through water erosion.
SRP researchers at the University of California, San Diego (UCSD) identified important mechanisms that plants use to respond to changes in the environment. Led by Julian Schroeder, Ph.D., they previously discovered proteins that make certain plants more tolerant to arsenic. Building on this and other findings, the research team is working to understand key mechanisms that function in heavy metal detoxification, transport, and accumulation in plants.
In a collaborative study between the University of Arizona and UCSD SRP Centers, researchers combined information about bacterial communities and plant genes in a new tool to identify key microbes important for plant establishment and survival. They plan to culture these microbes to study the mechanisms of plant-microbe-metal interactions.
Measuring Success of Bioremediation

At the University of North Carolina at Chapel Hill, previous SRP grantees identified relationships between incomplete PAH metabolism during bioremediation of soil and its toxicity. Led by Michael Aitken, Ph.D., they found that while a second-stage treatment using low levels of surfactants, which are commonly used as dispersing agents, may be a promising method to maximize PAH removal at hazardous waste sites, downstream effects should be considered. Even though this second-stage treatment removed PAHs, it also increased soil toxicity in many cases, emphasizing the need to better understand toxic by-products of PAH remediation.
Led by Staci Simonich, Ph.D., Oregon State University SRP grantees are also trying to understand PAH breakdown products in complex environmental mixtures at Superfund sites. They reported that even though steam-enhanced extraction effectively removed unsubstituted PAHs from contaminated soil, other, potentially more toxic, PAH derivatives are formed.
Small business grantee Microbial Insights develops tools using metabolomics to monitor changes to microbial communities during bioremediation at contaminated sites. Metabolomics measures all small organic molecules present in a biological sample that directly relate to active metabolism and can provide key insights into microbial community function and health. By studying microbial communities, researchers may be able to predict degradation stalls or development of potentially toxic byproducts.
At the University of California, Davis, researchers led by Tom Young, Ph.D., develop and evaluate approaches that combine analytic bioassay methods to assess toxicity reductions from bioremediation of Superfund sites. They aim to develop tools to optimize biodegradation of two contaminant mixtures, triazine herbicides and polycyclic aromatic hydrocarbons, which are representative of environmental exposures faced by the Yurok Tribe in Northern California, one of the Center's community partners.
The Microbiome
SRP researchers are identifying how the human gut microbiome interacts with contaminants and how changes to this microbiome may alter health outcomes after exposure to environmental chemicals.
Linking Exposures to Altered Microbiomes
Researchers at the Columbia University SRP Center identified associations between arsenic exposure and carotid intima-media thickness, which may play a role in development of atherosclerosis, and abundance of specific bacteria in the gut microbiome. According to the researchers, certain bacteria in the gut may contribute to the development of atherosclerosis, especially among individuals with higher levels of arsenic exposure.
Researchers at the University of California, Davis SRP Center found differences in the bacterial diversity of infants' fecal microbiomes when exposed to breast milk with and without detectable levels of the chemical triclosan. The researchers suggest some environmental chemicals, such as triclosan, may affect microbiome diversity.
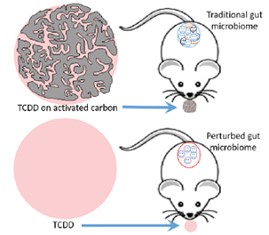
Michigan State University SRP Center researchers led by Syed Hashsham, Ph.D., discovered that the chemical 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) increases bacterial groups that harbor antibiotic resistance genes in the mouse microbiome. Their findings may explain the underlying mechanism for how TCDD can disrupt immune response and increase susceptibility to infectious disease. The group found that activated carbon reduces the effects of TCDD on microbes in the gut.
Identifying Differences in Microbiome Interactions
At the Dartmouth College SRP Center, scientists led by Margaret Karagas, Ph.D., explored differences in infants' gut microbiome based on arsenic exposure in a U.S. population. Compared to females, male infants experienced greater differences in their microbiomes with increasing arsenic exposure.
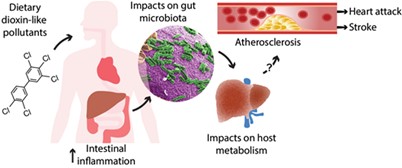
Researchers led by Bernhard Hennig, Ph.D., at the University of Kentucky explored the link between exposure to polychlorinated biphenyls (PCBs) and altered gut microbiomes in mice. Their findings suggest that PCB exposure may influence glucose and lipid metabolism by disrupting either bacterial fermentation processes or membrane integrity.
UCSD SRP Center researchers found an association between a metabolite in the blood that is derived from the gut microbiome and genes associated with both fatty liver disease and liver fibrosis that can be shared among families. According to the authors, the results suggest a link between genetics and a gut microbiome-derived metabolite in the heritability of non-alcoholic fatty liver disease.
to Top



