Features
SRP Researchers Develop Tools and Methods to Characterize the Exposome
The combined effects of contaminants from air, water, and food, as well as chemicals produced by the body, complicate efforts to find links between chemical exposures and disease. Because of this complexity, environmental health researchers are investigating the exposome, or the totality of exposures of an individual from conception onward.
Superfund Research Program (SRP) Center scientists and engineers are developing new tools and methods to better understand the exposome. These elements include exposure tools to describe the complex nature of the environment, methods to assess biological response from multiple stressors, and ways to integrate different types of data to characterize the effects of multiple exposures on health. A more comprehensive understanding of how the environment influences human health throughout the lifespan is an important step toward identifying and reducing disease risk.
Tools to Better Understand Complex Exposures

As the environmental health science field strives to better understand the complexity of chemical exposures, SRP researchers have developed non-targeted methods to understand and characterize a variety of compounds in the environment and in our bodies.
For example, after Hurricane Florence devastated parts of North and South Carolina in September 2018, Duke University SRP Center researchers Lee Ferguson, Ph.D., and Abigail Joyce, Ph.D., collected floodwater samples along the Neuse River and some of its tributaries. The Duke SRP Center's Analytical Chemistry Core is conducting non-targeted screening on the samples using high-resolution, accurate-mass mass spectrometry, a technique to identity the diverse set of compounds present in the sample.

With SRP and other NIEHS funding, Kim Anderson, Ph.D., and her team at Oregon State University (OSU) developed simple, wearable silicone wristbands to capture and measure a large number of diverse compounds at environmentally relevant concentrations. The wristbands have been used to study personal exposures following Hurricane Harvey, pesticide exposures in West Africa ,links between flame retardant exposure and social behaviors in children , and more.
OSU SRP Center researchers also have developed a method to automate non-targeted analysis of environmental samples using twoâdimensional gas chromatography coupled with time-of-flight mass spectrometry. Their method, which analyzes complex mixtures of comparative samples, reduces the potential for human error during heavy data handling and decreases data analysis time at least tenfold.
University of California (UC) Berkeley SRP Center researcher Stephen Rappaport, Ph.D., is using a battery of untargeted methods, notably metabolomics and proteomics, which are large-scale studies of metabolites and proteins, to characterize the exposome. In addition to characterizing exposures using dried blood spots , Rappaport also has shown that saliva may be a practical alternative to blood for characterizing such exposures. His work shows how saliva contains important molecular information and can be measured over time to construct individual exposure histories and to discover risk factors for disease.

At the Northeastern University SRP Center, researchers led by Roger Giese, Ph.D., developed a new cost-effective tool and accompanying method to extract substances from urine. The porous extraction paddle provides a simple method for solid-phase extraction of urine samples. This non-targeted technique can reveal large numbers of exposures to substances foreign to the body, or xenobiotics. By measuring the wide range of exposures in a non-targeted way and collecting subsequent health data, researchers may be able to find new associations between exposures and health outcomes.
Researchers led by Erin Baker, Ph.D., a North Carolina State University professor and Texas A&M University SRP Center researcher, characterized over 500 small molecules consisting of metabolites involved in biological pathways in the body and a variety of xenobiotics. Through this work, Baker and her team developed and shared a database of information about the small molecules, including structural information. According to the researchers, the information will greatly enable future metabolomic and exposomic studies by allowing others to more confidently identify these molecules and create better tools to thoroughly understand small molecule changes in biological and environmental systems.
Assessing Biological Response
In addition to improving what we know about how we are exposed to chemicals, researchers also are exploring new ways to assess how the body responds to exposures. By identifying more specific biological pathways activated in response to exposure, we may be able to better understand and characterize the cumulative impact of multiple environmental factors.
University of North Carolina at Chapel Hill SRP Center Director Rebecca Fry, Ph.D., is leading a team to identify metabolite changes and proteome shifts associated with arsenic exposure. Assessing changes in the proteome and metabolome may provide insight into how prenatal arsenic exposure could affect important pathways responsible for maintaining normal cell processes in the body.
As part of the Northeastern University SRP Center, researchers led by John Meeker, Ph.D., are identifying markers of oxidative stress and inflammation associated with exposure to phenols and parabens. They also are developing a method to identify environmental chemical mixtures affecting health outcomes.
University of Washington SRP Center researchers led by Clem Furlong, Ph.D., have developed a method using dried blood spots to measure an individual's activity levels of paraoxaonase-1 (PON1), an enzyme that can detoxify organophosphorus insecticides. Measuring PON1 status as a marker of susceptibility helps to identify mothers and children who are highly sensitive to exposure to organophosphorus insecticides.
Integrating Data to Characterize the Exposome
Developing new models that combine data on exposures and effects of chemicals is becoming an important part of characterizing the exposome. SRP researchers are developing frameworks to understand and organize these data.
Led by UC Berkeley SRP Center Director Martyn Smith, Ph.D., researchers have developed an intrinsic (I) by extrinsic (E) (I x E) concept to incorporate the multi-factorial and dynamic nature of health and disease. Intrinsic vulnerability or resilience includes a variety of biological factors, including genetic traits, while extrinsic factors comprise all biologically relevant external stressors encountered across the lifespan. According to the authors, this approach provides a unified, conceptual basis for integrating results from multiple areas of research, including genomics, genes by environment, developmental origins of health and disease, and the exposome. Smith also uses key characteristics as a basis for organizing data on mechanisms that may lead to cancer and uses exposomics to study interactions between chronic stress and environmental chemicals that disrupt stress response pathways.
Researchers at the Boston University SRP Center led by Thomas Webster, Ph.D., are using novel statistical models to evaluate the effects of chemical mixtures and nonchemical stressors on health. They have developed and assessed ageneralized concentration addition model to predict the joint effects of environmental chemicals that affect the aryl hydrocarbon receptor, a protein required for organisms to develop properly.
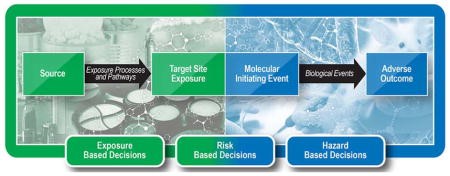
Oregon State SRP Center researchers led by Justin Teeguarden, Ph.D., have developed a framework to organize exposure data into aggregate exposure pathways (AEP) to provide critical information about how environmental concentrations can be related to human exposures. Looking ahead, the researchers envision direct linkages between AEPs and adverse outcome pathways to more efficiently integrate exposure assessment and hazard identification.
to Top



